"sodium channel, voltage-gated, type III, beta"
ZFIN 
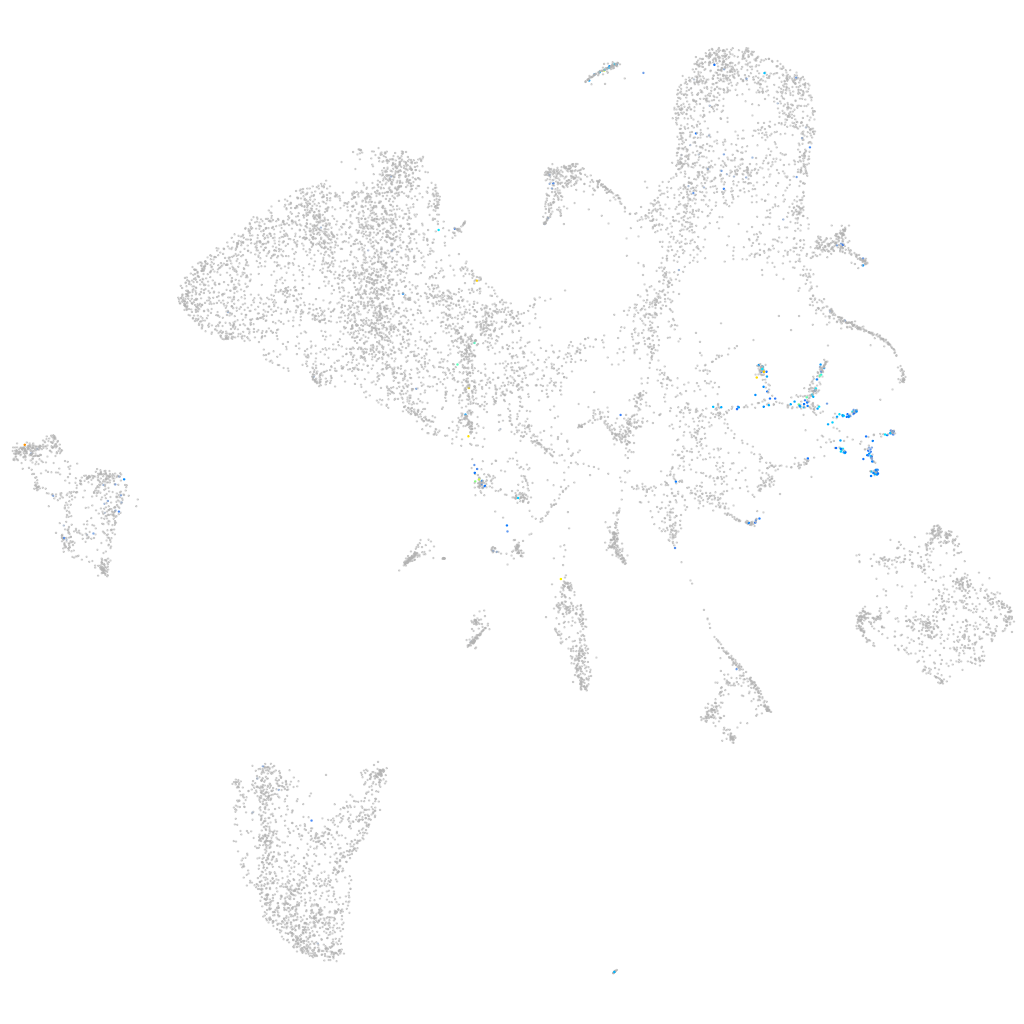
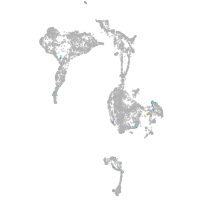
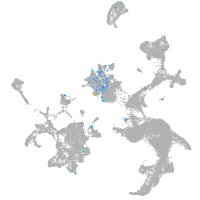
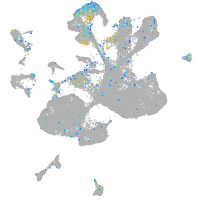
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.397 | ahcy | -0.112 |
| scg3 | 0.385 | gapdh | -0.109 |
| neurod1 | 0.370 | hdlbpa | -0.106 |
| pcsk1nl | 0.361 | gamt | -0.103 |
| mir7a-1 | 0.353 | nupr1b | -0.095 |
| pax6b | 0.346 | rpl6 | -0.095 |
| isl1 | 0.335 | aldob | -0.095 |
| ndufa4l2a | 0.331 | gatm | -0.093 |
| kcnj11 | 0.330 | gstr | -0.093 |
| pcsk1 | 0.327 | aldh7a1 | -0.093 |
| si:dkey-153k10.9 | 0.323 | mat1a | -0.090 |
| insm1a | 0.311 | fbp1b | -0.090 |
| tspan7b | 0.304 | gstt1a | -0.089 |
| syt1a | 0.301 | adka | -0.088 |
| gcgb | 0.301 | cx32.3 | -0.088 |
| vat1 | 0.296 | bhmt | -0.087 |
| SLC5A10 | 0.295 | rps20 | -0.085 |
| rims2a | 0.293 | aqp12 | -0.085 |
| slc30a2 | 0.288 | aldh6a1 | -0.084 |
| scg2b | 0.288 | eif4ebp1 | -0.083 |
| scg2a | 0.287 | apoa4b.1 | -0.083 |
| pcsk2 | 0.285 | glud1b | -0.082 |
| cacna1c | 0.285 | dhdhl | -0.081 |
| rprmb | 0.280 | fabp3 | -0.080 |
| atp6v0cb | 0.278 | hspd1 | -0.080 |
| ptn | 0.277 | aldh1l1 | -0.080 |
| abhd15a | 0.276 | gstp1 | -0.079 |
| LOC100537384 | 0.275 | apoc2 | -0.079 |
| id4 | 0.274 | hspe1 | -0.078 |
| ins | 0.273 | gcshb | -0.078 |
| foxo1a | 0.272 | si:dkey-16p21.8 | -0.078 |
| mir375-2 | 0.270 | phyhd1 | -0.078 |
| rnasekb | 0.268 | tktb | -0.077 |
| abcc8 | 0.265 | ppa1b | -0.077 |
| aldocb | 0.261 | apoc1 | -0.077 |