"secretagogin, EF-hand calcium binding protein"
ZFIN 
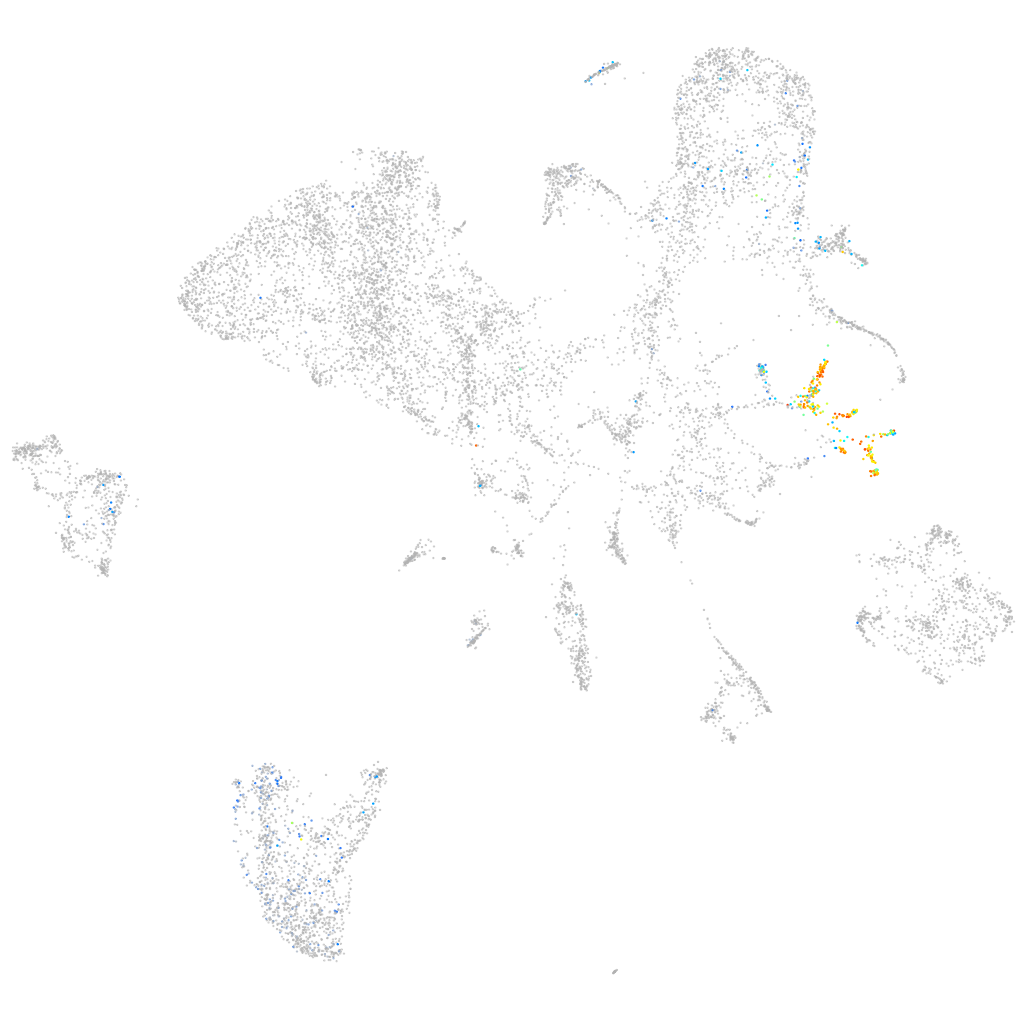
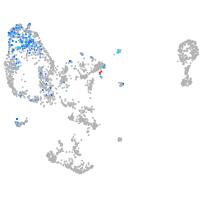
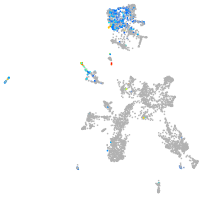
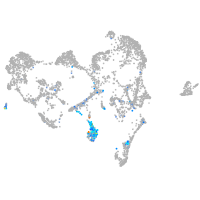
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcsk2 | 0.696 | ahcy | -0.186 |
| scg3 | 0.651 | gapdh | -0.180 |
| rprmb | 0.640 | gamt | -0.165 |
| isl1 | 0.633 | gstp1 | -0.151 |
| si:dkey-153k10.9 | 0.632 | fabp3 | -0.150 |
| kcnj11 | 0.628 | aldob | -0.150 |
| arxa | 0.621 | apoc2 | -0.148 |
| neurod1 | 0.604 | apoa1b | -0.147 |
| pax6b | 0.594 | si:dkey-16p21.8 | -0.147 |
| rims2a | 0.562 | apoa4b.1 | -0.146 |
| rasd4 | 0.556 | bhmt | -0.146 |
| pcsk1nl | 0.555 | aldh6a1 | -0.145 |
| gcga | 0.553 | gstr | -0.145 |
| SLC5A10 | 0.544 | gstt1a | -0.145 |
| scg2a | 0.544 | gatm | -0.144 |
| syt1a | 0.539 | apoc1 | -0.144 |
| ndufa4l2a | 0.527 | hdlbpa | -0.142 |
| c2cd4a | 0.518 | rps20 | -0.142 |
| tspan7b | 0.512 | afp4 | -0.141 |
| pcsk1 | 0.507 | fbp1b | -0.141 |
| vat1 | 0.499 | eno3 | -0.139 |
| mir7a-1 | 0.495 | mat1a | -0.136 |
| abhd15a | 0.489 | mgst1.2 | -0.134 |
| LOC100537384 | 0.486 | shmt1 | -0.132 |
| dkk3b | 0.479 | glud1b | -0.131 |
| fam49bb | 0.476 | cx32.3 | -0.130 |
| camk1da | 0.467 | tfa | -0.130 |
| ptn | 0.465 | agxta | -0.130 |
| gcgb | 0.463 | apoa2 | -0.130 |
| gpc1a | 0.461 | ppa1b | -0.129 |
| insm1a | 0.459 | gnmt | -0.127 |
| gpr142 | 0.453 | aldh7a1 | -0.127 |
| pyyb | 0.450 | wu:fj16a03 | -0.127 |
| foxo1a | 0.450 | rpl6 | -0.126 |
| abcc8 | 0.450 | apobb.1 | -0.125 |