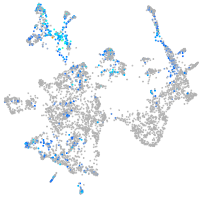
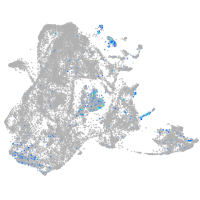
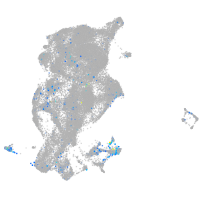
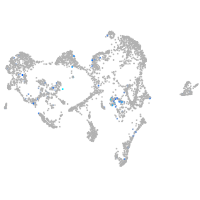
secretogranin III
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| myl10 | 0.618 | syncrip | -0.480 |
| neurod1 | 0.615 | seta | -0.450 |
| si:dkey-208c12.2 | 0.577 | ddx4 | -0.428 |
| ppm1bb | 0.550 | ctsla | -0.409 |
| tmem167b | 0.547 | stmn1a | -0.406 |
| insm1a | 0.545 | anp32b | -0.403 |
| cldnh | 0.544 | h1m | -0.399 |
| mnx1 | 0.541 | nasp | -0.387 |
| nkx6.2 | 0.541 | rpl6 | -0.379 |
| LOC103910736 | 0.541 | hnrnph1l | -0.378 |
| gng2 | 0.540 | gra | -0.370 |
| myt1b | 0.534 | ranbp1 | -0.367 |
| eml2 | 0.531 | mki67 | -0.365 |
| ins | 0.527 | zgc:56699 | -0.362 |
| col9a1b | 0.526 | ccna2 | -0.360 |
| rtn1a | 0.520 | hnrnpub | -0.360 |
| LOC101883836 | 0.520 | hdgfl2 | -0.359 |
| arfip2a | 0.504 | sf3b2 | -0.359 |
| myl9a | 0.503 | safb | -0.357 |
| atcaya | 0.496 | srrm1 | -0.352 |
| CT573204.1 | 0.495 | NC-002333.4 | -0.349 |
| ntd5 | 0.493 | zmat2 | -0.347 |
| rsph9 | 0.492 | rpl7l1 | -0.345 |
| map6a | 0.489 | hnrnpa1a | -0.345 |
| s100a11 | 0.481 | baz1b | -0.345 |
| ptmaa | 0.479 | dnd1 | -0.341 |
| cplx2l | 0.477 | ilf3b | -0.337 |
| ift52 | 0.462 | etf1b | -0.337 |
| slc26a6 | 0.461 | smc1al | -0.334 |
| pax6b | 0.461 | ppig | -0.334 |
| tp53inp2 | 0.460 | ebna1bp2 | -0.334 |
| gapdhs | 0.456 | matr3l1.1 | -0.333 |
| mdkb | 0.454 | cd2bp2 | -0.333 |
| ece1 | 0.451 | cx43.4 | -0.332 |
| stxbp1a | 0.450 | morf4l1 | -0.331 |