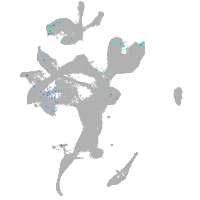
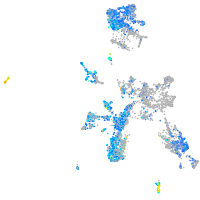
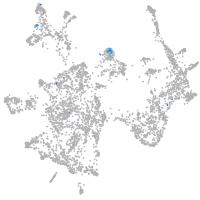
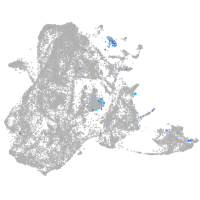
secretogranin II (chromogranin C) a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.544 | ahcy | -0.153 |
| scg3 | 0.528 | gapdh | -0.152 |
| si:dkey-153k10.9 | 0.509 | gamt | -0.146 |
| pcsk2 | 0.489 | aldob | -0.131 |
| SLC5A10 | 0.486 | gatm | -0.128 |
| pcsk1nl | 0.484 | bhmt | -0.124 |
| neurod1 | 0.474 | nupr1b | -0.122 |
| syt1a | 0.467 | mat1a | -0.121 |
| ndufa4l2a | 0.449 | fbp1b | -0.121 |
| pcsk1 | 0.444 | rps20 | -0.121 |
| pax6b | 0.433 | gstr | -0.119 |
| isl1 | 0.428 | apoa1b | -0.117 |
| mir7a-1 | 0.425 | gstt1a | -0.116 |
| gpc1a | 0.415 | si:dkey-16p21.8 | -0.115 |
| kcnj11 | 0.414 | apoa4b.1 | -0.115 |
| rims2a | 0.408 | apoc2 | -0.115 |
| vamp2 | 0.402 | aldh6a1 | -0.114 |
| insm1a | 0.401 | hdlbpa | -0.114 |
| dkk3b | 0.389 | eno3 | -0.112 |
| camk1da | 0.388 | fabp3 | -0.110 |
| cpe | 0.387 | apoc1 | -0.110 |
| scg2b | 0.382 | aqp12 | -0.109 |
| vat1 | 0.376 | afp4 | -0.109 |
| zgc:101731 | 0.375 | eif4ebp1 | -0.108 |
| rprmb | 0.372 | cx32.3 | -0.107 |
| fam49bb | 0.369 | gnmt | -0.105 |
| foxo1a | 0.367 | agxtb | -0.104 |
| tspan7b | 0.366 | shmt1 | -0.104 |
| c2cd4a | 0.365 | aldh7a1 | -0.104 |
| ptn | 0.362 | agxta | -0.103 |
| egr4 | 0.358 | hspe1 | -0.102 |
| aldocb | 0.357 | apoa2 | -0.101 |
| slc30a2 | 0.355 | adka | -0.101 |
| lysmd2 | 0.354 | gcshb | -0.101 |
| ins | 0.353 | scp2a | -0.100 |