"scavenger receptor class B, member 2a"
ZFIN 
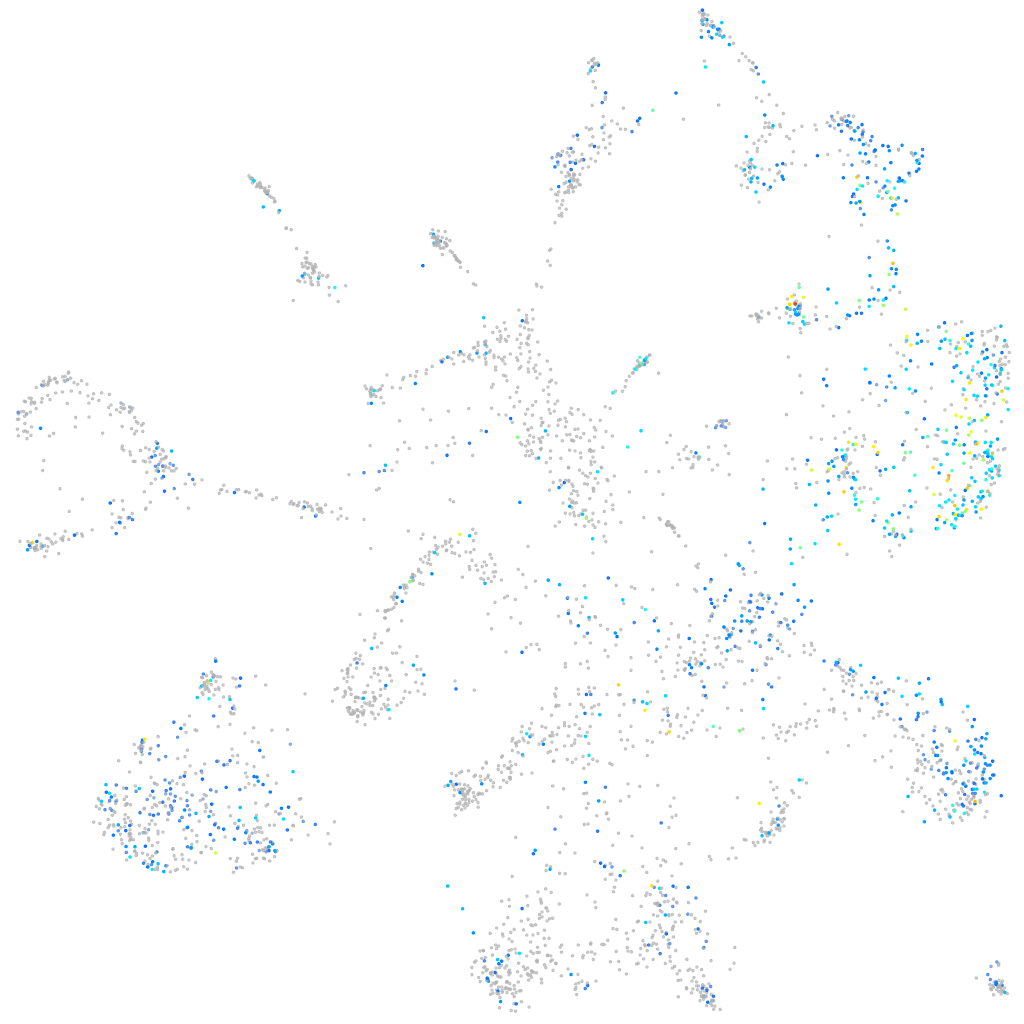
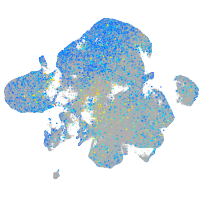
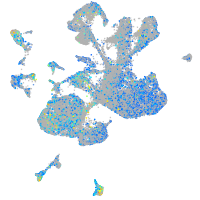
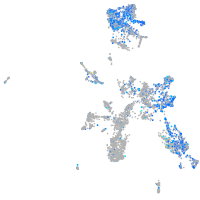
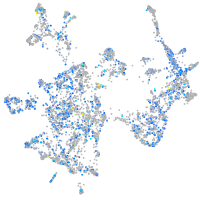
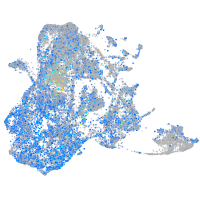
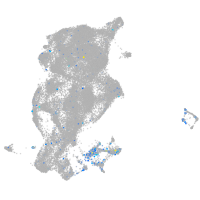
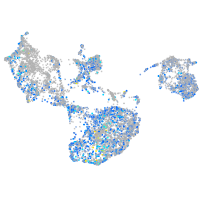
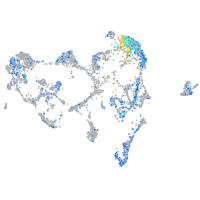
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| podxl | 0.455 | BX088707.3 | -0.217 |
| jam2b | 0.450 | tpm1 | -0.208 |
| bcam | 0.354 | acta2 | -0.207 |
| aqp1a.1 | 0.340 | tagln | -0.205 |
| cd151 | 0.325 | myh11a | -0.188 |
| cav1 | 0.324 | ptmaa | -0.184 |
| krt94 | 0.314 | fhl2a | -0.167 |
| cavin2b | 0.312 | mylkb | -0.161 |
| rhag | 0.308 | myl9a | -0.159 |
| ftr82 | 0.307 | lmod1b | -0.158 |
| colec11 | 0.303 | tpm2 | -0.150 |
| tmem88b | 0.300 | mylka | -0.141 |
| cavin1b | 0.290 | csrp1b | -0.140 |
| aqp8a.1 | 0.290 | gapdhs | -0.137 |
| txn | 0.284 | cald1b | -0.134 |
| COLEC10 | 0.279 | cnn1b | -0.131 |
| si:ch211-156j16.1 | 0.278 | fhl1a | -0.122 |
| cdon | 0.277 | ak1 | -0.122 |
| tmem88a | 0.260 | inka1a | -0.121 |
| phactr4a | 0.258 | desmb | -0.117 |
| postnb | 0.251 | cox4i2 | -0.117 |
| krt15 | 0.245 | ppp1cbl | -0.117 |
| bnc2 | 0.244 | myl6 | -0.116 |
| cdh2 | 0.243 | myocd | -0.115 |
| dag1 | 0.242 | calm2a | -0.112 |
| myrf | 0.242 | fxyd1 | -0.111 |
| lrrc15 | 0.241 | sat1a.2 | -0.110 |
| crb2a | 0.240 | rbm24a | -0.108 |
| akap12b | 0.240 | si:ch211-270g19.5 | -0.108 |
| hspb1 | 0.240 | wnt11r | -0.107 |
| cx43.4 | 0.237 | mgll | -0.107 |
| krt8 | 0.236 | eno3 | -0.107 |
| ehd1b | 0.231 | igfbp7 | -0.106 |
| arl4aa | 0.228 | lbh | -0.105 |
| myh10 | 0.228 | BX323087.1 | -0.103 |