SREBF chaperone
ZFIN 
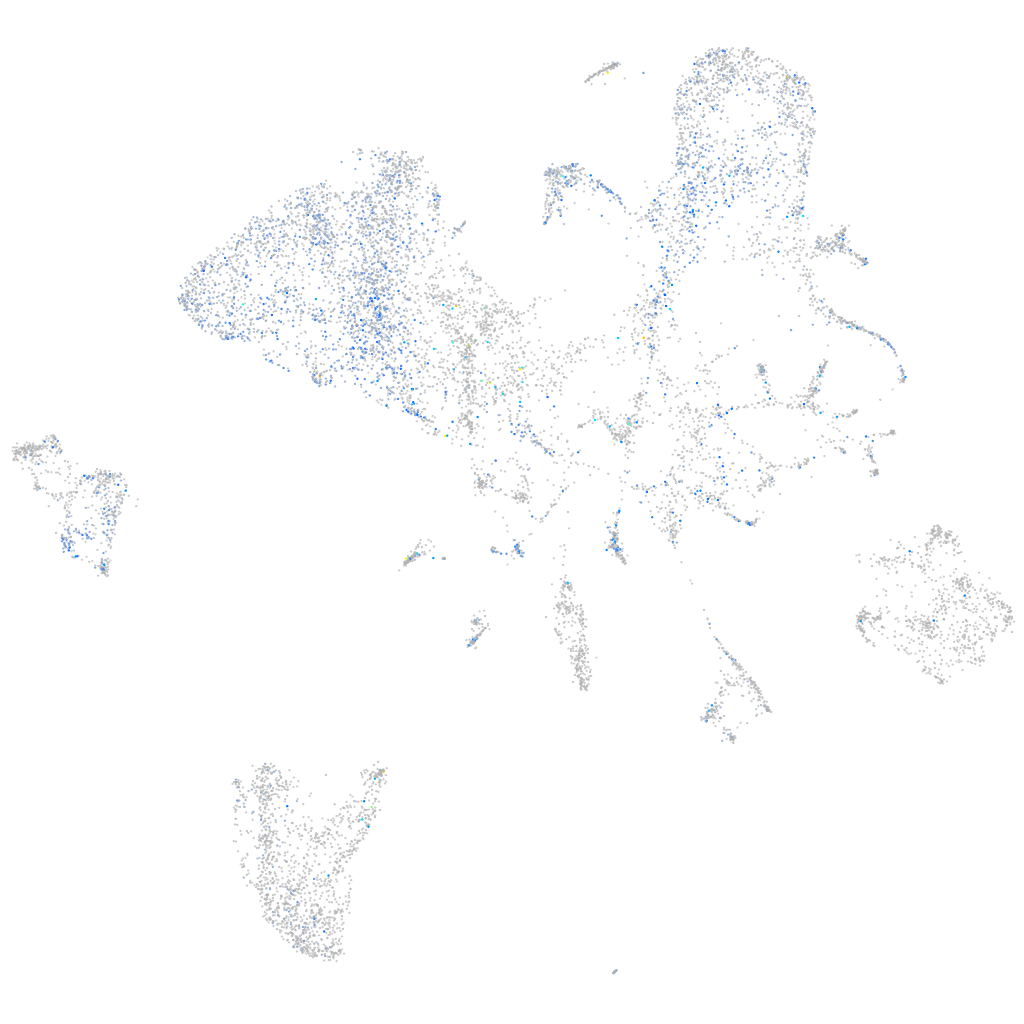
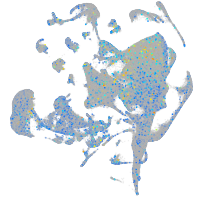
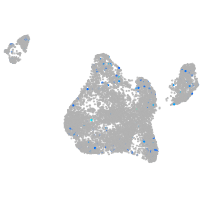
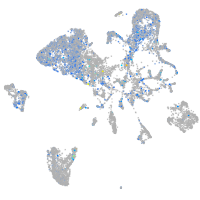
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mt-nd1 | 0.206 | rtn1a | -0.110 |
| mt-atp6 | 0.196 | prss59.1 | -0.103 |
| slco1d1 | 0.194 | ctrl | -0.103 |
| ugt1a7 | 0.187 | ctrb1 | -0.102 |
| hadhab | 0.180 | ela3l | -0.102 |
| hspd1 | 0.177 | ela2l | -0.102 |
| atp5f1b | 0.176 | marcksl1b | -0.101 |
| cyp8b1 | 0.173 | CELA1 (1 of many) | -0.101 |
| tecrb | 0.173 | zgc:112160 | -0.101 |
| mttp | 0.173 | prss59.2 | -0.101 |
| srd5a2a | 0.173 | ela2 | -0.101 |
| aco1 | 0.171 | cpa5 | -0.100 |
| aldh1l1 | 0.170 | prss1 | -0.100 |
| mt-nd2 | 0.170 | pdia2 | -0.099 |
| pgrmc1 | 0.169 | c6ast4 | -0.099 |
| atp5mc1 | 0.166 | si:ch211-240l19.5 | -0.099 |
| hacd2 | 0.165 | cel.1 | -0.099 |
| rtn4a | 0.165 | fep15 | -0.099 |
| lipf | 0.164 | erp27 | -0.099 |
| sdr16c5b | 0.163 | cpb1 | -0.098 |
| hadhb | 0.163 | zgc:136461 | -0.098 |
| eno3 | 0.160 | sycn.2 | -0.098 |
| chchd10 | 0.160 | apoda.2 | -0.098 |
| fdx1 | 0.160 | hspb1 | -0.097 |
| mt-cyb | 0.159 | cel.2 | -0.095 |
| mt-nd4 | 0.158 | endou | -0.095 |
| cldn15lb | 0.157 | akap12b | -0.095 |
| cox6b1 | 0.157 | zgc:92137 | -0.094 |
| msrb2 | 0.156 | amy2a | -0.093 |
| pgam1a | 0.156 | marcksb | -0.093 |
| sod1 | 0.156 | cpa4 | -0.093 |
| prdx2 | 0.156 | cpa1 | -0.092 |
| f5 | 0.156 | si:dkey-14d8.6 | -0.091 |
| gstk1 | 0.156 | si:ch211-240l19.6 | -0.091 |
| uqcrc1 | 0.155 | tmed6 | -0.091 |