"spermidine/spermine N1-acetyltransferase 1a, duplicate 2"
ZFIN 
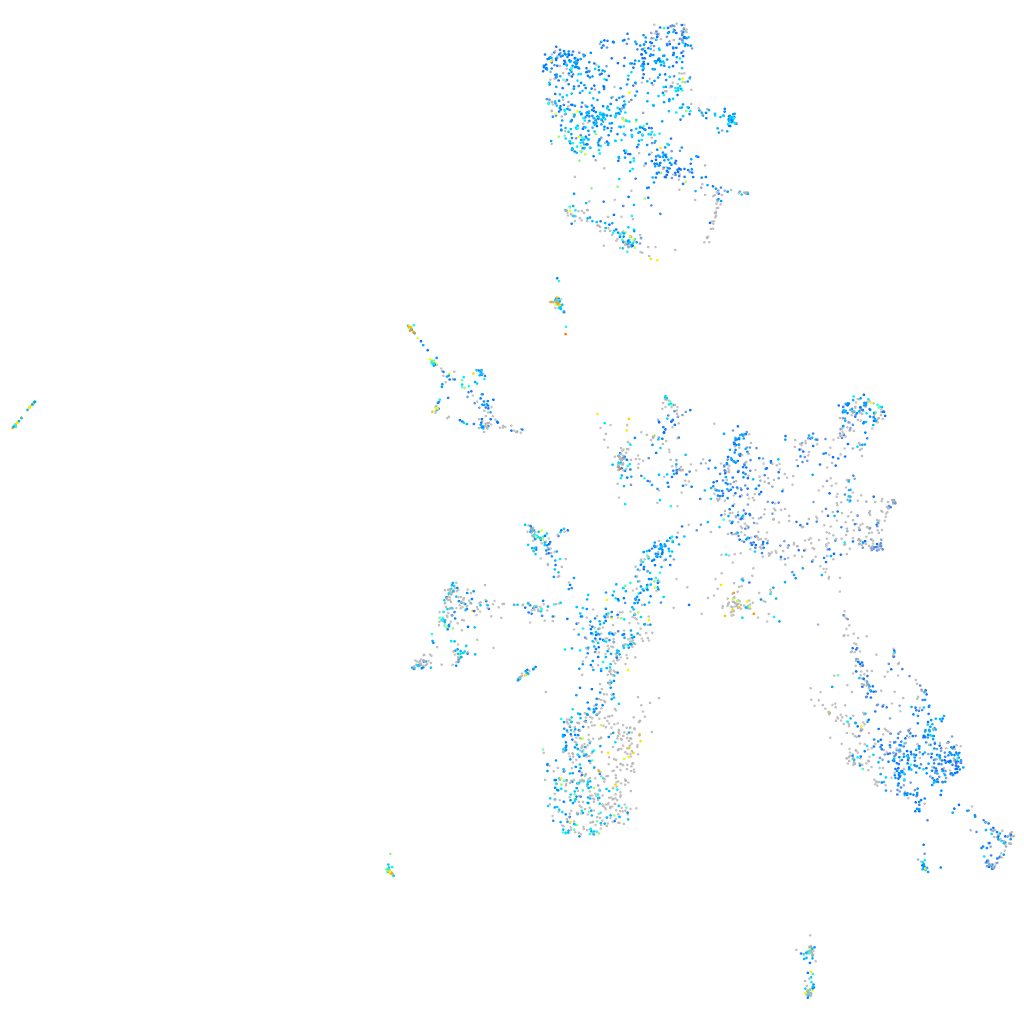
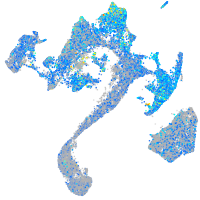
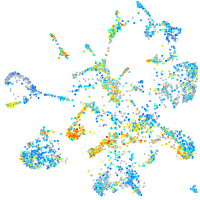
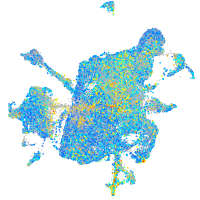
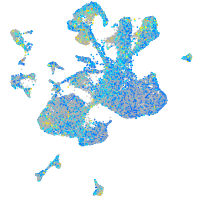
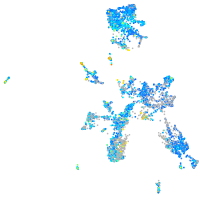
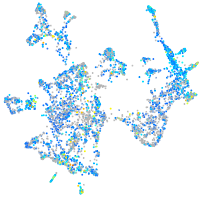
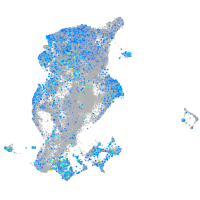
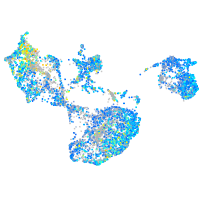
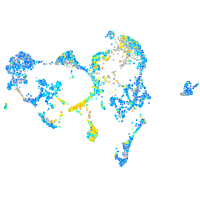
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ucp2 | 0.282 | NC-002333.4 | -0.179 |
| scg3 | 0.270 | sp8b | -0.165 |
| prox1a | 0.258 | sp8a | -0.163 |
| nr4a1 | 0.250 | her12 | -0.160 |
| dusp5 | 0.247 | otx1 | -0.159 |
| mcl1a | 0.247 | si:ch211-243g18.2 | -0.156 |
| jun | 0.238 | dlx5a | -0.154 |
| tob1b | 0.236 | her2 | -0.151 |
| XLOC-013964 | 0.231 | XLOC-032526 | -0.145 |
| bhlhe40 | 0.228 | her4.1 | -0.143 |
| foxq1a | 0.226 | sox21a | -0.142 |
| nfasca | 0.226 | dut | -0.138 |
| calm1b | 0.225 | chaf1a | -0.137 |
| tob1a | 0.223 | hells | -0.136 |
| scgn | 0.223 | tmsb4x | -0.134 |
| rasd1 | 0.222 | fezf1 | -0.133 |
| prelid3b | 0.222 | nasp | -0.131 |
| egr4 | 0.220 | zgc:174938 | -0.130 |
| si:ch211-56a11.2 | 0.220 | dlx6a | -0.128 |
| hepacam2 | 0.218 | krt18b | -0.127 |
| higd1a | 0.217 | cdca7b | -0.125 |
| vill | 0.217 | lig1 | -0.125 |
| scg2a | 0.211 | nid2a | -0.125 |
| cst3 | 0.211 | rac1a | -0.124 |
| cldnh | 0.211 | fen1 | -0.124 |
| dusp2 | 0.208 | cks1b | -0.124 |
| egr2a | 0.207 | BX927258.1 | -0.124 |
| CR936442.1 | 0.206 | dkc1 | -0.124 |
| mir375-2 | 0.205 | dlx3b | -0.123 |
| xbp1 | 0.204 | gmnn | -0.122 |
| hopx | 0.204 | pcna | -0.122 |
| syngr2b | 0.203 | si:dkey-261h17.1 | -0.121 |
| calm1a | 0.202 | shmt1 | -0.120 |
| si:ch211-196l7.4 | 0.201 | rpa2 | -0.119 |
| atf4b | 0.200 | ppp1r3b | -0.119 |