SAM and SH3 domain containing 1a
ZFIN 
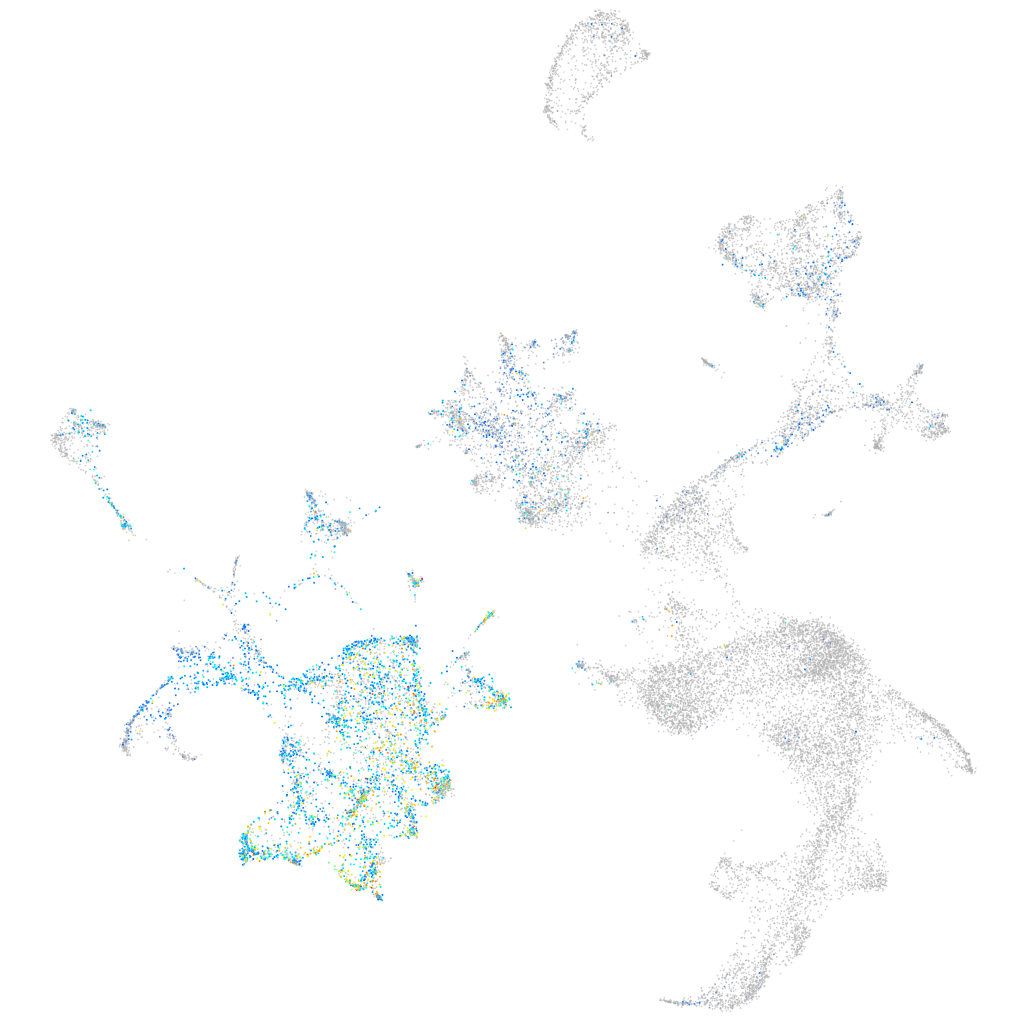
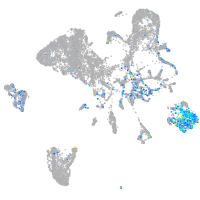
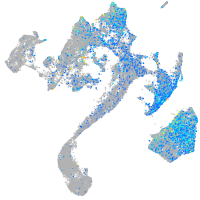
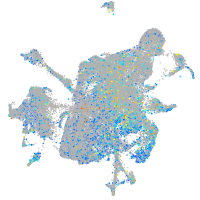
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdh5 | 0.518 | prdx2 | -0.367 |
| krt18a.1 | 0.506 | hbae1.1 | -0.348 |
| clec14a | 0.483 | hbae1.3 | -0.337 |
| si:ch211-156j16.1 | 0.476 | hbbe2 | -0.334 |
| sox7 | 0.472 | hbae3 | -0.333 |
| krt8 | 0.472 | hbbe1.3 | -0.328 |
| cldn5b | 0.467 | hbbe1.1 | -0.319 |
| she | 0.467 | cahz | -0.318 |
| tpm4a | 0.466 | blvrb | -0.307 |
| egfl7 | 0.466 | hemgn | -0.300 |
| akap12b | 0.461 | hbbe1.2 | -0.292 |
| si:ch211-248e11.2 | 0.459 | fth1a | -0.290 |
| kdrl | 0.451 | alas2 | -0.280 |
| si:dkeyp-97a10.2 | 0.445 | slc4a1a | -0.270 |
| cpn1 | 0.445 | tspo | -0.269 |
| pecam1 | 0.442 | nt5c2l1 | -0.259 |
| myct1a | 0.441 | creg1 | -0.259 |
| tie1 | 0.441 | si:ch211-250g4.3 | -0.258 |
| mcamb | 0.440 | zgc:163057 | -0.251 |
| kdr | 0.438 | epb41b | -0.248 |
| plk2b | 0.436 | gpx1a | -0.245 |
| si:dkey-261h17.1 | 0.433 | si:ch211-207c6.2 | -0.234 |
| plvapb | 0.429 | nmt1b | -0.228 |
| podxl | 0.429 | plac8l1 | -0.222 |
| etv2 | 0.427 | tmod4 | -0.217 |
| si:ch211-145b13.6 | 0.426 | hbae5 | -0.198 |
| sox18 | 0.425 | vdac2 | -0.195 |
| igfbp7 | 0.424 | tmem14ca | -0.194 |
| col4a1 | 0.422 | sptb | -0.190 |
| arhgap31 | 0.419 | tfr1a | -0.189 |
| ecscr | 0.419 | uros | -0.185 |
| tspan4b | 0.416 | rfesd | -0.183 |
| cd81a | 0.414 | klf1 | -0.182 |
| ramp2 | 0.407 | si:ch211-227m13.1 | -0.180 |
| fam174b | 0.406 | add2 | -0.179 |