sterile alpha motif domain containing 13
ZFIN 
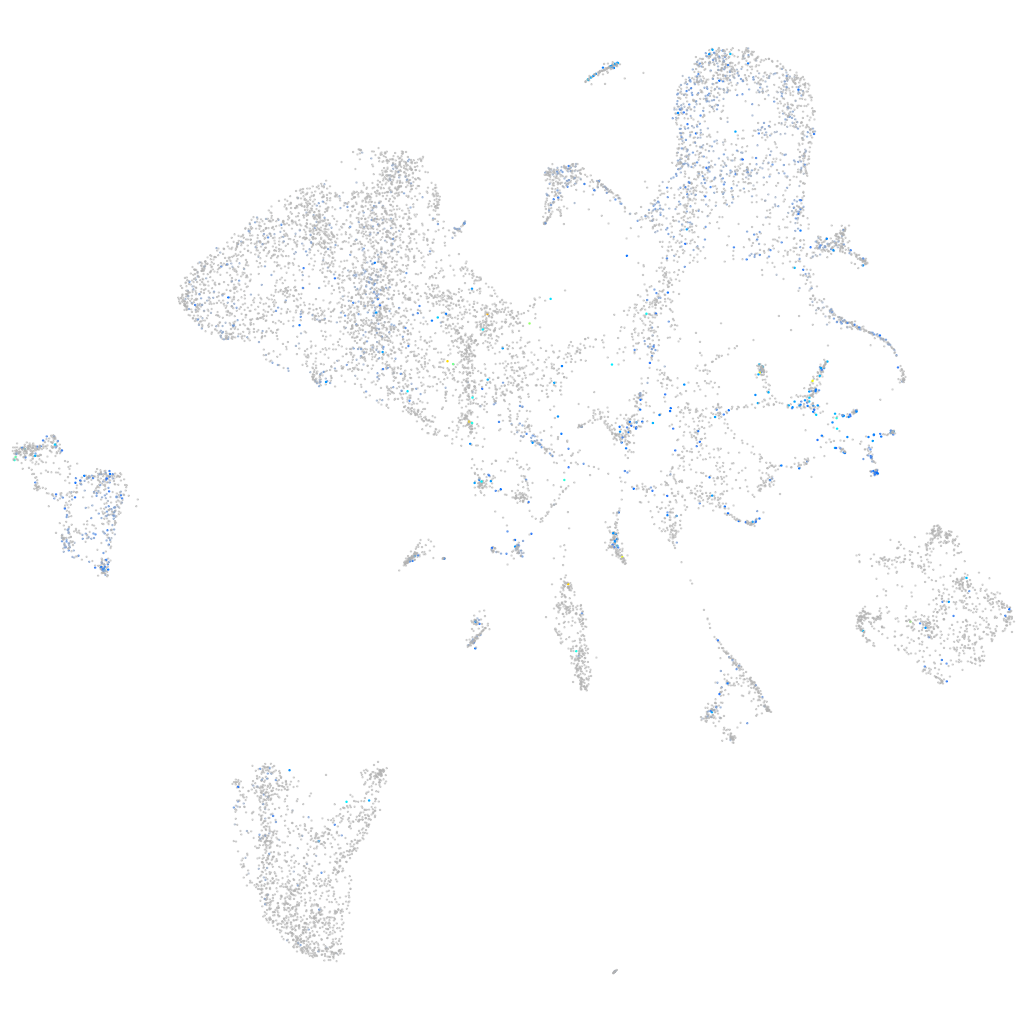
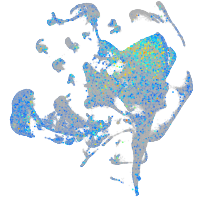
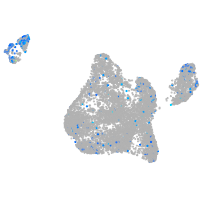
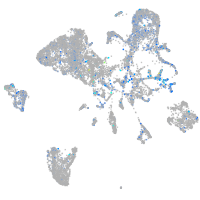
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.174 | bhmt | -0.126 |
| neurod1 | 0.168 | apoc1 | -0.114 |
| si:ch1073-429i10.3.1 | 0.154 | aqp12 | -0.113 |
| gapdhs | 0.154 | agxtb | -0.109 |
| syt1a | 0.152 | gamt | -0.107 |
| pcsk1nl | 0.152 | gatm | -0.103 |
| calm3b | 0.152 | fetub | -0.095 |
| mir7a-1 | 0.151 | eif4ebp3 | -0.095 |
| scgn | 0.149 | wu:fj16a03 | -0.094 |
| calm2b | 0.148 | mat1a | -0.091 |
| camk1da | 0.148 | tfa | -0.091 |
| calm1b | 0.147 | ppdpfa | -0.089 |
| si:dkey-153k10.9 | 0.147 | apoa2 | -0.088 |
| arpc3 | 0.147 | gc | -0.088 |
| calml4a | 0.146 | pnp4b | -0.087 |
| gpc1a | 0.145 | fabp10a | -0.087 |
| kcnj11 | 0.145 | apom | -0.087 |
| ppiab | 0.144 | ces2 | -0.086 |
| insm1a | 0.144 | serpina1 | -0.086 |
| c2cd4a | 0.143 | rbp4 | -0.085 |
| vat1 | 0.143 | uox | -0.084 |
| h3f3a | 0.143 | fgb | -0.084 |
| rnasekb | 0.143 | zgc:123103 | -0.083 |
| mir375-2 | 0.142 | kng1 | -0.083 |
| arxa | 0.141 | rbp2b | -0.082 |
| tango2 | 0.141 | ambp | -0.082 |
| scg2a | 0.141 | hao1 | -0.082 |
| kcnn3 | 0.140 | serpina1l | -0.082 |
| zgc:101731 | 0.138 | ttr | -0.081 |
| tmed7 | 0.137 | LOC110437731 | -0.081 |
| GCA | 0.137 | f2 | -0.081 |
| gnb1a | 0.136 | zgc:136461 | -0.081 |
| tspan7b | 0.136 | hpda | -0.081 |
| pax6b | 0.136 | sycn.2 | -0.081 |
| sat1a.2 | 0.136 | ela2 | -0.080 |