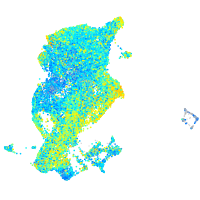
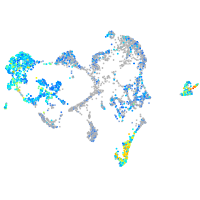
S100 calcium binding protein A10b
ZFIN 
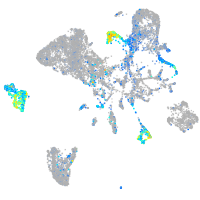
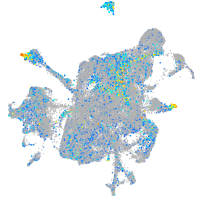
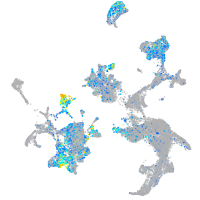
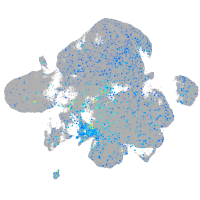
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| krt4 | 0.713 | elavl3 | -0.419 |
| cfl1l | 0.682 | rtn1a | -0.386 |
| aqp3a | 0.661 | insm1a | -0.381 |
| zgc:158343 | 0.658 | tmeff1b | -0.360 |
| s100v1 | 0.654 | stmn1b | -0.333 |
| lgals3b | 0.654 | atp6v0cb | -0.330 |
| krt18a.1 | 0.642 | hmgb3a | -0.330 |
| pycard | 0.642 | epb41a | -0.325 |
| malb | 0.640 | ebf3a | -0.308 |
| tmsb1 | 0.638 | sox11a | -0.306 |
| sult6b1 | 0.634 | nova2 | -0.301 |
| capns1a | 0.631 | insm1b | -0.300 |
| dnase1l4.1 | 0.625 | NC-002333.4 | -0.297 |
| krt8 | 0.616 | si:dkeyp-75h12.5 | -0.291 |
| cldne | 0.613 | zgc:65894 | -0.288 |
| sparc | 0.605 | vdac3 | -0.286 |
| si:ch211-195b11.3 | 0.597 | si:dkey-276j7.1 | -0.286 |
| krt97 | 0.591 | sncb | -0.283 |
| s100v2 | 0.582 | tuba1a | -0.280 |
| elf3 | 0.577 | myt1a | -0.279 |
| gsta.1 | 0.568 | gng3 | -0.279 |
| agr1 | 0.565 | tspan2a | -0.278 |
| cldnb | 0.560 | si:dkey-56m19.5 | -0.277 |
| si:dkey-87o1.2 | 0.558 | rnasekb | -0.273 |
| tmem176l.4 | 0.555 | zc4h2 | -0.272 |
| ahnak | 0.555 | si:ch211-133n4.4 | -0.264 |
| zgc:153284 | 0.548 | CU467822.1 | -0.259 |
| eef1da | 0.548 | CR383676.1 | -0.257 |
| si:ch73-204p21.2 | 0.542 | neurod1 | -0.257 |
| zgc:153911 | 0.539 | atp2b1a | -0.256 |
| LOC799574 | 0.538 | hnrnpa0a | -0.255 |
| cnn2 | 0.535 | LOC100537342 | -0.250 |
| phlda2 | 0.533 | gnao1a | -0.250 |
| ftr83 | 0.533 | atp2b1b | -0.249 |
| cd9b | 0.532 | h3f3d | -0.248 |