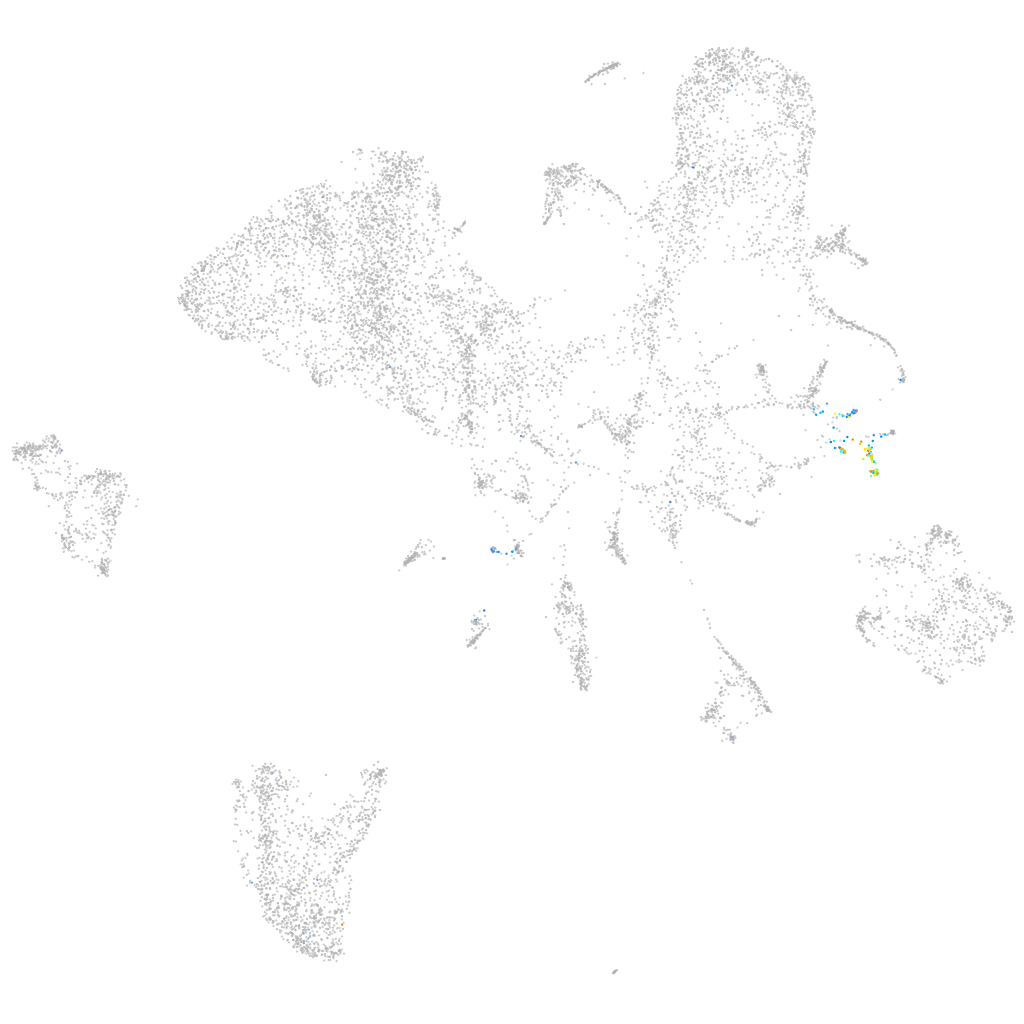
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm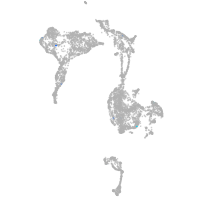 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 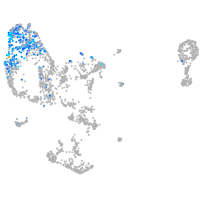 |
neural |
spinal cord / glia |
eye |
taste / olfactory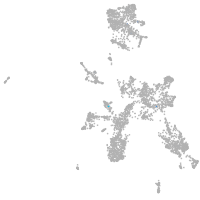 |
otic / lateral line |
epidermis |
periderm |
fin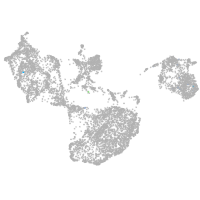 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ndufa4l2a | 0.805 | ahcy | -0.118 |
| ins | 0.717 | gapdh | -0.115 |
| ptn | 0.650 | aldob | -0.115 |
| slc30a2 | 0.641 | gamt | -0.099 |
| ucn3l | 0.548 | gstp1 | -0.098 |
| scgn | 0.544 | rps20 | -0.096 |
| sst1.1 | 0.522 | eno3 | -0.096 |
| pcsk2 | 0.517 | gatm | -0.092 |
| pcsk1 | 0.487 | fbp1b | -0.091 |
| scg2a | 0.486 | hdlbpa | -0.090 |
| si:dkey-153k10.9 | 0.483 | tuba8l2 | -0.089 |
| rims2a | 0.476 | glud1b | -0.087 |
| kcnj11 | 0.475 | gstt1a | -0.086 |
| kcnip3b | 0.462 | bhmt | -0.085 |
| cntnap2a | 0.460 | nupr1b | -0.084 |
| cadm2a | 0.431 | apoc2 | -0.084 |
| g6pcb | 0.429 | apoa4b.1 | -0.084 |
| renbp | 0.427 | apoa1b | -0.083 |
| dkk3b | 0.414 | si:dkey-16p21.8 | -0.083 |
| camk1da | 0.410 | gsta.1 | -0.082 |
| tmem163b | 0.408 | gstr | -0.082 |
| abhd15a | 0.407 | aldh9a1a.1 | -0.082 |
| hdac9b | 0.406 | hspe1 | -0.080 |
| rprmb | 0.399 | sdr16c5b | -0.080 |
| pax6b | 0.396 | cox6b1 | -0.080 |
| scg2b | 0.395 | afp4 | -0.080 |
| zgc:194990 | 0.394 | fabp2 | -0.079 |
| fam49bb | 0.394 | gpx4a | -0.079 |
| cd164l2 | 0.393 | lgals2b | -0.078 |
| pcsk1nl | 0.389 | aldh6a1 | -0.078 |
| kcnj19a | 0.389 | suclg1 | -0.078 |
| prokr1a | 0.388 | mat1a | -0.078 |
| rims2b | 0.386 | cx32.3 | -0.077 |
| gpr22a | 0.385 | gcshb | -0.077 |
| neurod1 | 0.380 | gnmt | -0.077 |