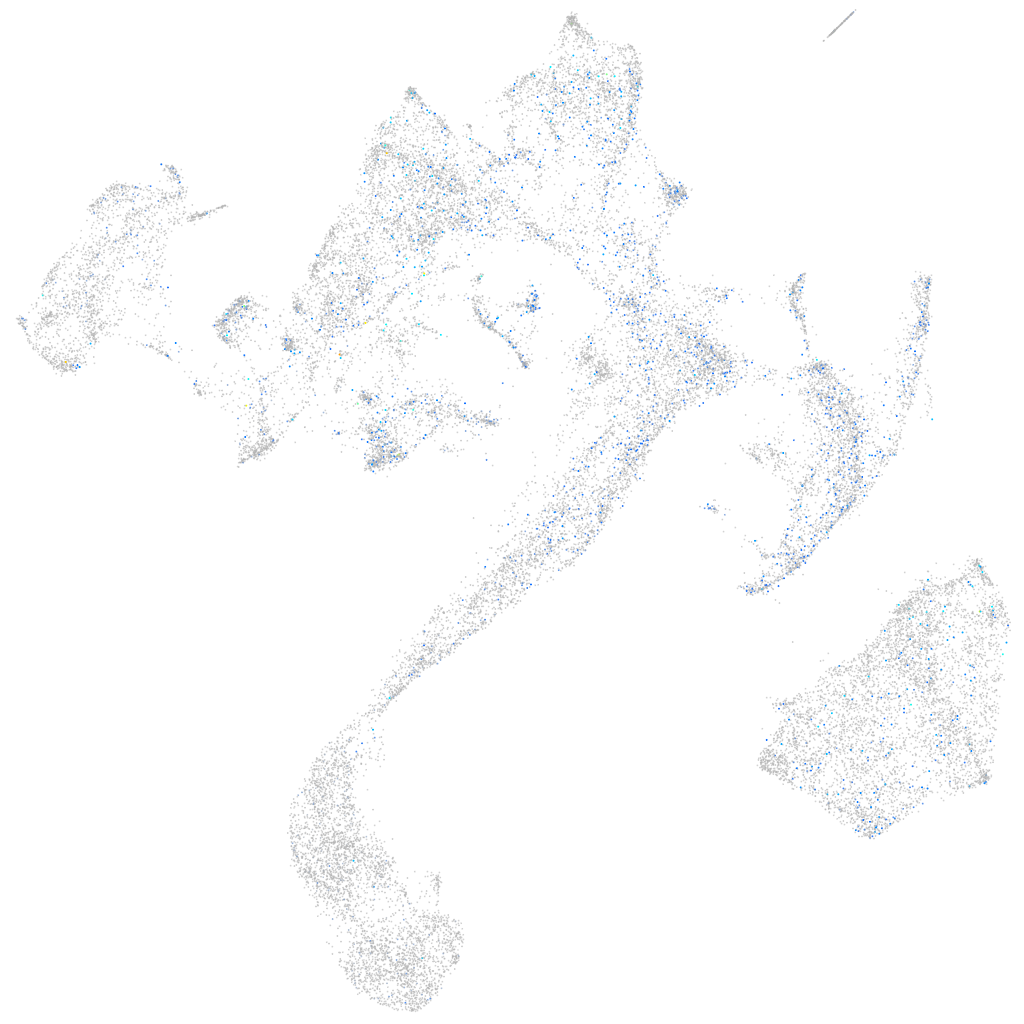
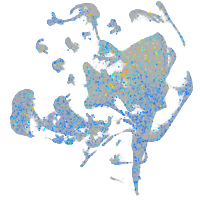
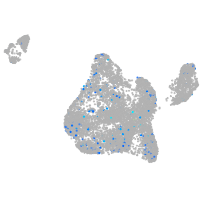
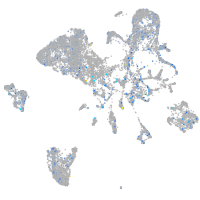
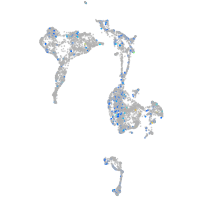
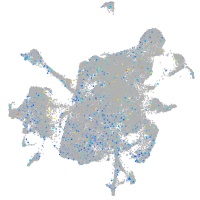
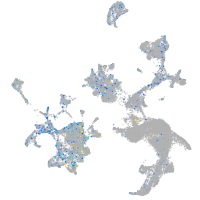
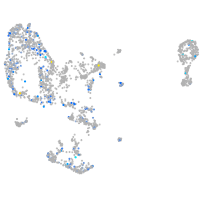
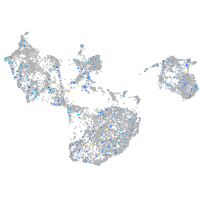
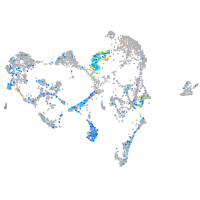
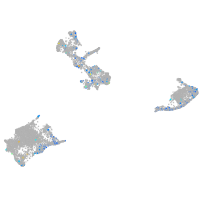
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| actb2 | 0.085 | ckmb | -0.076 |
| ubc | 0.081 | ckma | -0.076 |
| hsp90ab1 | 0.079 | atp2a1 | -0.072 |
| h3f3a | 0.078 | si:ch73-367p23.2 | -0.072 |
| hnrnpa0l | 0.077 | ak1 | -0.072 |
| kcnj12a | 0.076 | actn3a | -0.072 |
| h3f3d | 0.074 | mylz3 | -0.071 |
| cfl1 | 0.074 | acta1b | -0.070 |
| hspa8 | 0.073 | gapdh | -0.070 |
| ppib | 0.072 | ldb3b | -0.070 |
| actb1 | 0.072 | tnnc2 | -0.070 |
| eef2b | 0.071 | tnnt3a | -0.070 |
| tnfrsf13b | 0.070 | tpma | -0.069 |
| CT573423.1 | 0.070 | tmod4 | -0.069 |
| rorc | 0.070 | actc1b | -0.069 |
| CR383676.1 | 0.069 | mylpfa | -0.069 |
| zgc:153867 | 0.069 | myom1a | -0.068 |
| ssr3 | 0.069 | XLOC-025819 | -0.067 |
| ran | 0.069 | aldoab | -0.067 |
| chrnb3b | 0.069 | smyd1a | -0.067 |
| COX7A2 (1 of many) | 0.068 | ank1a | -0.067 |
| mfap2 | 0.068 | CABZ01078594.1 | -0.067 |
| zgc:56493 | 0.068 | neb | -0.067 |
| si:dkey-261h17.1 | 0.067 | tnni2a.4 | -0.067 |
| lrp1ab | 0.067 | XLOC-005350 | -0.067 |
| cirbpb | 0.067 | pvalb2 | -0.066 |
| marcksl1a | 0.066 | si:ch211-255p10.3 | -0.066 |
| znf997 | 0.066 | pvalb1 | -0.066 |
| mdka | 0.066 | myoz1b | -0.066 |
| CABZ01034596.1 | 0.066 | XLOC-006515 | -0.065 |
| h2afvb | 0.066 | mylpfb | -0.065 |
| fthl27 | 0.065 | prr33 | -0.065 |
| eef1a1l1 | 0.065 | pgam2 | -0.065 |
| FO904898.6 | 0.065 | tnnt3b | -0.065 |
| hmgn2 | 0.065 | XLOC-001975 | -0.065 |