RUNX1 partner transcriptional co-repressor 1
ZFIN 
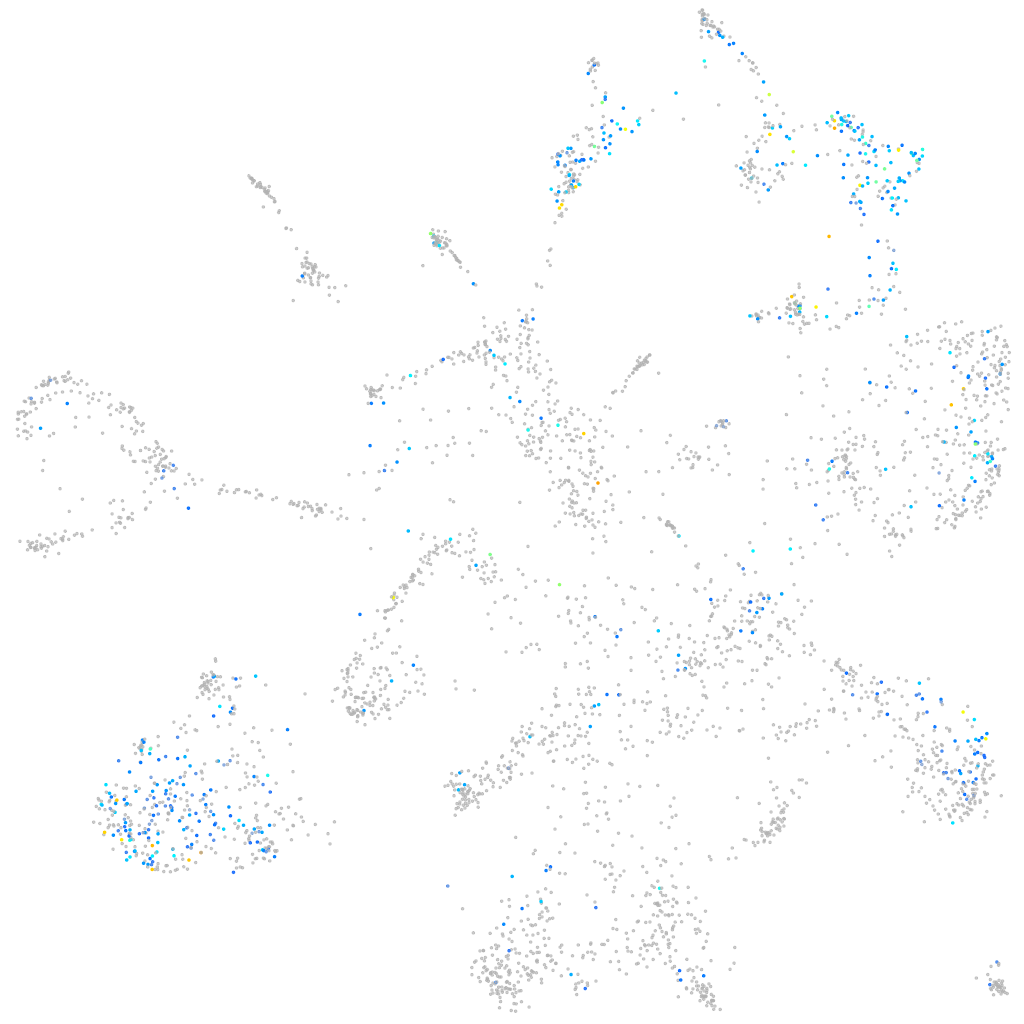
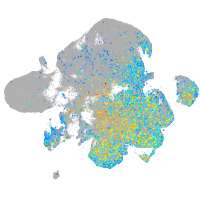
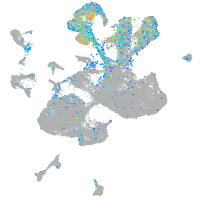
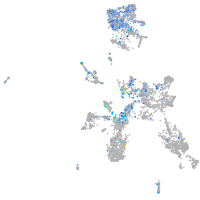
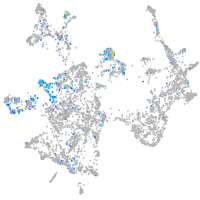
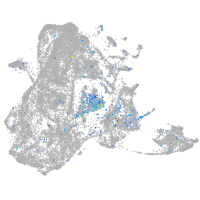
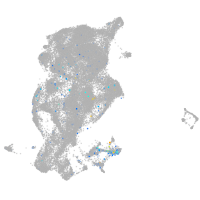
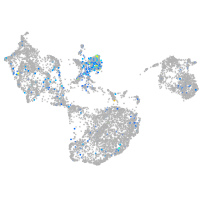
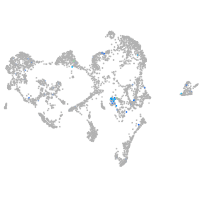
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gata5 | 0.277 | tpm1 | -0.149 |
| sfrp5 | 0.261 | pmp22a | -0.148 |
| tnni1b | 0.254 | BX088707.3 | -0.146 |
| alpi.1 | 0.253 | XLOC-041870 | -0.128 |
| mmel1 | 0.249 | acta2 | -0.113 |
| lmod2a | 0.248 | col4a1 | -0.112 |
| krt94 | 0.239 | igfbp7 | -0.108 |
| synpo2lb | 0.238 | ak1 | -0.108 |
| tnnt2a | 0.214 | tagln | -0.107 |
| gata6 | 0.208 | foxf1 | -0.106 |
| si:ch211-214p16.2 | 0.206 | mylkb | -0.100 |
| shisa2b | 0.205 | itga1 | -0.098 |
| angpt1 | 0.201 | inhbaa | -0.097 |
| nova2 | 0.201 | myl9a | -0.095 |
| COLEC10 | 0.195 | ptch2 | -0.094 |
| tbx5a | 0.195 | aoc2 | -0.093 |
| krt8 | 0.192 | col1a2 | -0.089 |
| angptl6 | 0.192 | tpm2 | -0.088 |
| XLOC-042222 | 0.191 | nkx2.3 | -0.088 |
| tmem88b | 0.190 | angptl5 | -0.085 |
| cxadr | 0.185 | XLOC-025423 | -0.084 |
| FITM1 (1 of many) | 0.183 | rbpms2a | -0.083 |
| alx4a | 0.182 | zgc:153704 | -0.082 |
| nr0b2a | 0.179 | tmem119b | -0.081 |
| CABZ01075068.1 | 0.179 | col11a1a | -0.081 |
| arvcfb | 0.177 | myh11a | -0.081 |
| atp2a2a | 0.177 | desmb | -0.081 |
| ncs1a | 0.175 | iscub | -0.081 |
| nexn | 0.174 | mamdc2a | -0.080 |
| podxl | 0.174 | lmod1b | -0.079 |
| ildr2 | 0.173 | inka1a | -0.079 |
| clec19a | 0.173 | col6a2 | -0.078 |
| elavl3 | 0.170 | csrp1b | -0.078 |
| fabp11a | 0.170 | col4a6 | -0.077 |
| krt18a.1 | 0.169 | cald1b | -0.076 |