RUN and FYVE domain containing 2
ZFIN 
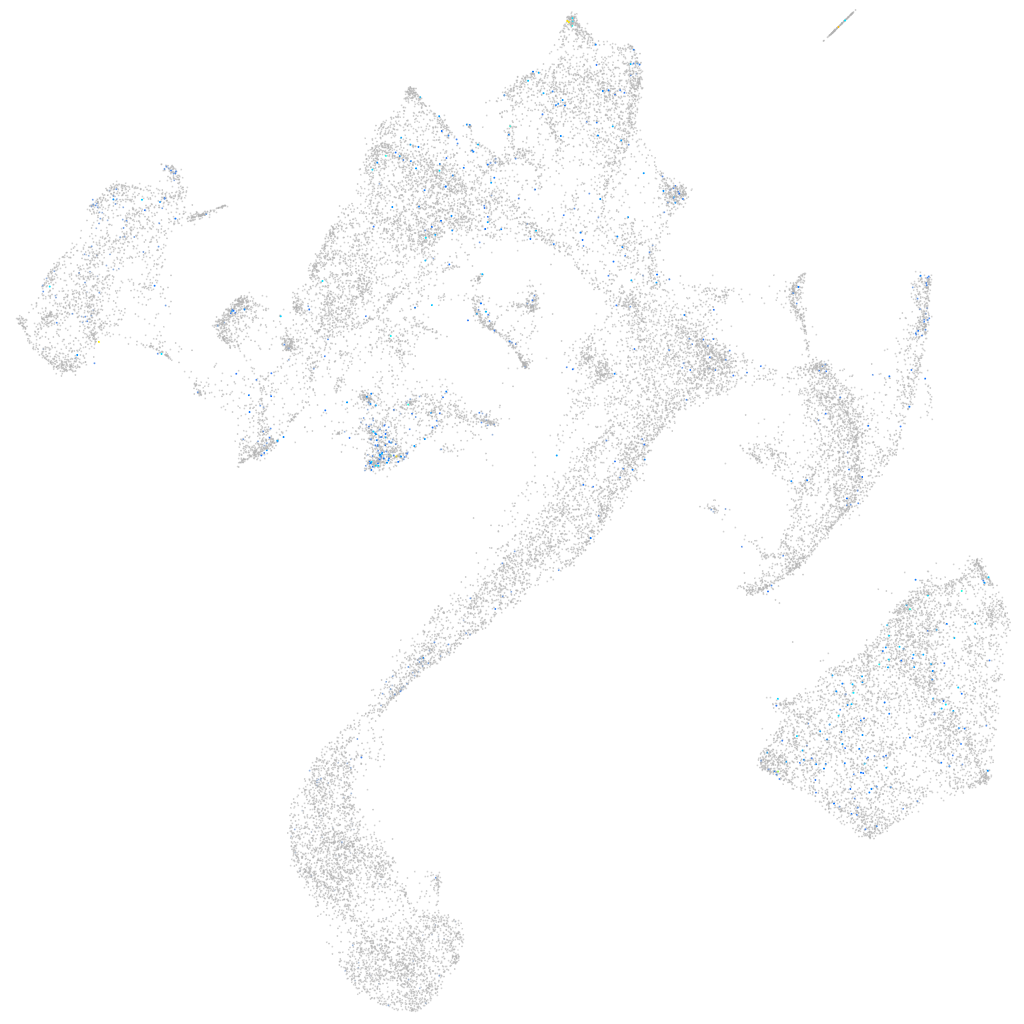
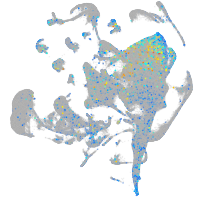
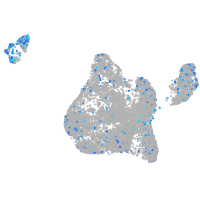
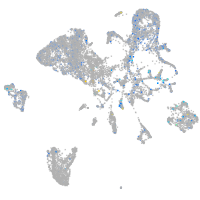
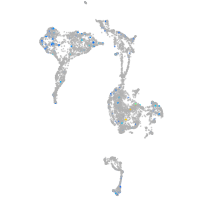
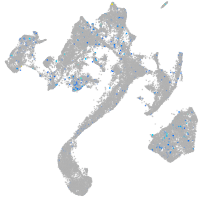
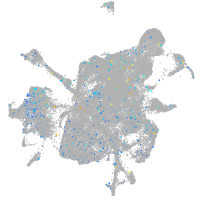
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR788316.4 | 0.099 | gatm | -0.042 |
| CABZ01085700.1 | 0.098 | si:dkey-151g10.6 | -0.041 |
| LOC101882164 | 0.097 | ryr3 | -0.037 |
| olfm1b | 0.096 | rps7 | -0.036 |
| nsg2 | 0.093 | rplp0 | -0.036 |
| rtn1b | 0.092 | myl1 | -0.035 |
| stmn1b | 0.092 | tma16 | -0.035 |
| elavl3 | 0.090 | myoz1b | -0.035 |
| maptb | 0.088 | casq1a | -0.035 |
| adcyap1b | 0.086 | mylpfb | -0.035 |
| zgc:101840 | 0.085 | CABZ01061524.1 | -0.035 |
| id4 | 0.084 | si:ch211-255p10.3 | -0.035 |
| gpm6aa | 0.083 | slc25a4 | -0.035 |
| myt1la | 0.083 | myom2a | -0.034 |
| sprn | 0.083 | tnni2a.4 | -0.033 |
| ephx4 | 0.081 | pvalb1 | -0.033 |
| jagn1a | 0.080 | pvalb2 | -0.033 |
| vamp2 | 0.080 | atp2a1l | -0.033 |
| rnasekb | 0.079 | mylz3 | -0.032 |
| gng3 | 0.079 | c1qbp | -0.032 |
| tuba1c | 0.078 | ampd1 | -0.032 |
| scg2b | 0.078 | vdac2 | -0.032 |
| cspg5a | 0.078 | tnnt3b | -0.032 |
| stmn2a | 0.077 | rps27a | -0.031 |
| cdk5r2a | 0.077 | eef1b2 | -0.031 |
| sncb | 0.077 | myhz1.3 | -0.030 |
| stx1b | 0.076 | tpma | -0.030 |
| si:dkey-56f14.7 | 0.075 | atp5mc3b | -0.030 |
| eno2 | 0.075 | atp1a2a | -0.030 |
| ribc1 | 0.075 | gamt | -0.030 |
| si:ch211-10a23.2 | 0.074 | hrc | -0.030 |
| SMIM18 | 0.074 | si:ch73-367p23.2 | -0.030 |
| gpr85 | 0.074 | rps13 | -0.030 |
| grin2bb | 0.074 | ahcy | -0.030 |
| BX927275.2 | 0.073 | hsc70 | -0.030 |