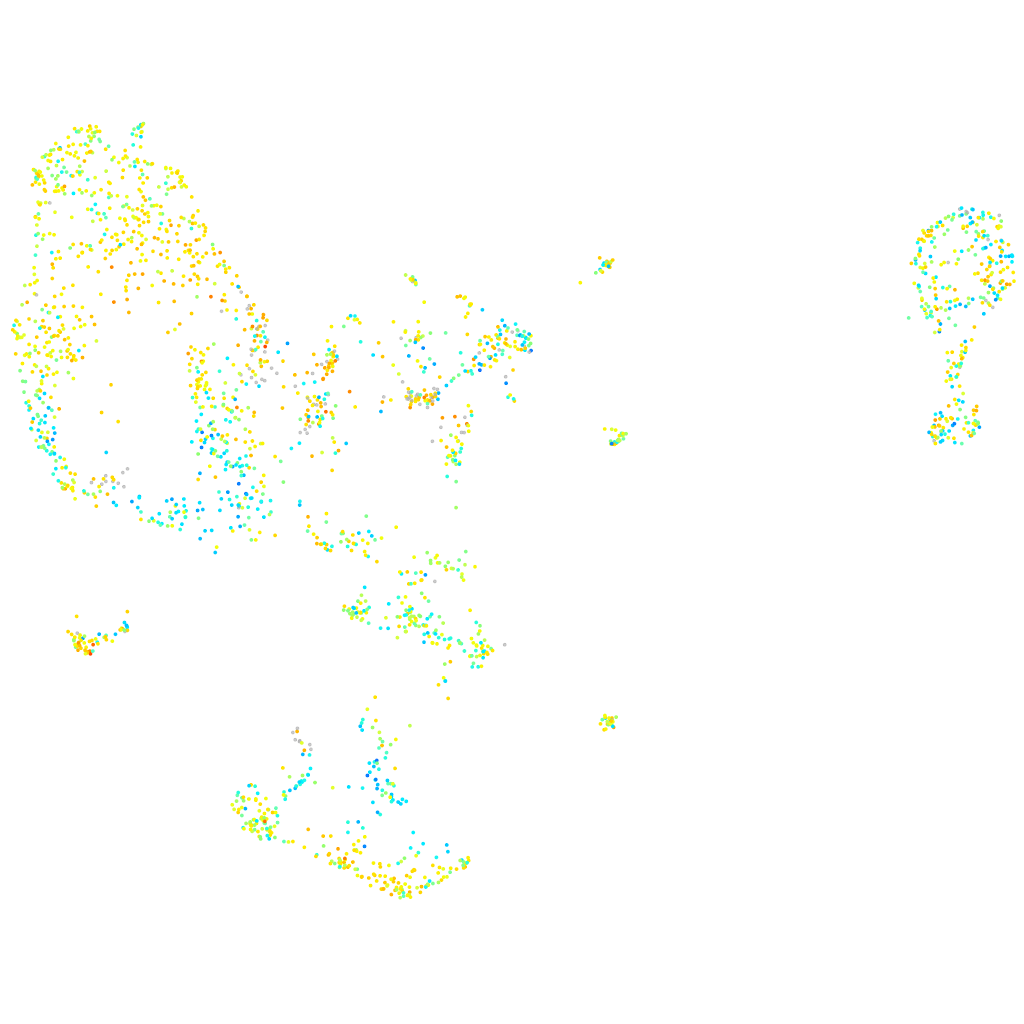"ribosomal protein S27, isoform 2"
ZFIN 
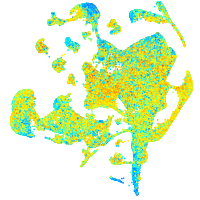
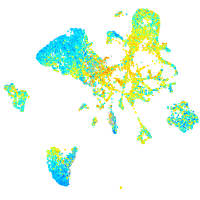
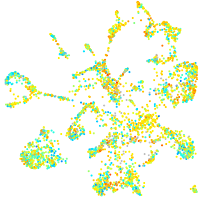
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rpl18a | 0.302 | cldn17 | -0.189 |
| rps26l | 0.301 | icn2 | -0.146 |
| rpl13a | 0.298 | cnmd | -0.134 |
| rpl17 | 0.293 | CU467822.1 | -0.130 |
| rpl15 | 0.289 | gpr158b | -0.125 |
| rps8a | 0.288 | inavaa | -0.122 |
| rpl5a | 0.283 | tmem74b | -0.122 |
| nme2b.1 | 0.283 | hbae1.1 | -0.120 |
| rpl34 | 0.279 | acta1b | -0.119 |
| rpl31 | 0.279 | XLOC-017089 | -0.119 |
| rps16 | 0.274 | otos | -0.117 |
| rack1 | 0.269 | cntnap5b | -0.116 |
| rpl14 | 0.269 | tuba1c | -0.113 |
| rpl9 | 0.267 | nsfa | -0.113 |
| rpl35a | 0.263 | hbbe3 | -0.111 |
| rps15 | 0.258 | cyt1 | -0.111 |
| rps19 | 0.258 | si:ch211-255p10.3 | -0.110 |
| naca | 0.257 | rimkla | -0.110 |
| rpl23a | 0.246 | nbeaa | -0.107 |
| rps13 | 0.246 | krt4 | -0.107 |
| rpl12 | 0.244 | hbae1.3 | -0.106 |
| rps9 | 0.236 | lingo1a | -0.106 |
| rps5 | 0.235 | rap1gap2a | -0.105 |
| adh5 | 0.234 | hbbe1.3 | -0.105 |
| eef1b2 | 0.225 | scg3 | -0.105 |
| mibp2 | 0.224 | BX571952.1 | -0.105 |
| sod1 | 0.222 | fam168a | -0.105 |
| rpl10 | 0.222 | CT583728.23 | -0.105 |
| rps3a | 0.222 | bricd5 | -0.105 |
| eef1da | 0.222 | dclk2b | -0.104 |
| rpl30 | 0.222 | ttc39c | -0.104 |
| rpl18 | 0.221 | CABZ01043955.1 | -0.104 |
| ndufa4l | 0.221 | LOC101883320 | -0.104 |
| gstp1 | 0.220 | her4.3 | -0.104 |
| rps3 | 0.219 | cyt1l | -0.103 |