regulation of nuclear pre-mRNA domain containing 2a
ZFIN 
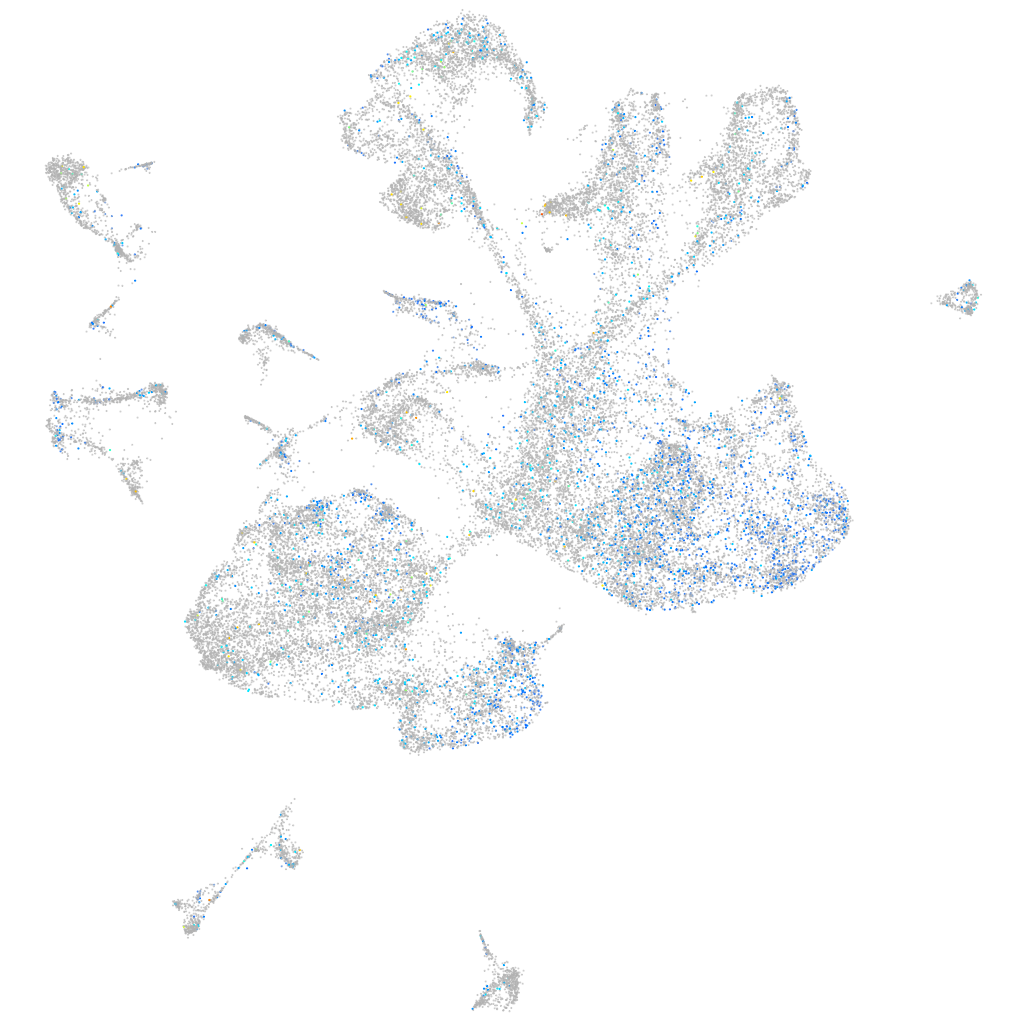
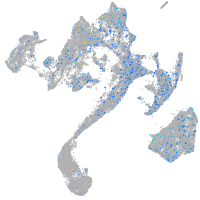
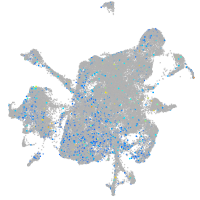
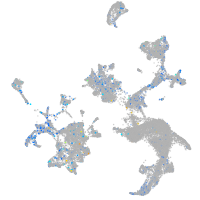
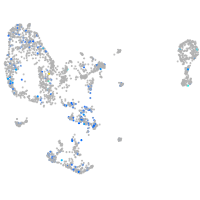
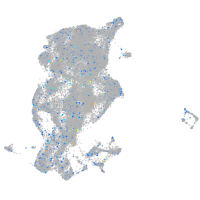
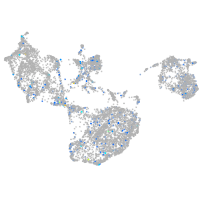
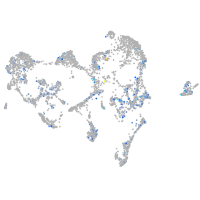
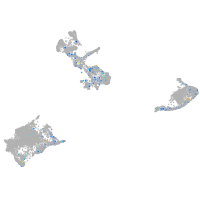
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ranbp1 | 0.089 | ckbb | -0.058 |
| hmga1a | 0.088 | pvalb1 | -0.056 |
| hmgb2b | 0.088 | pvalb2 | -0.056 |
| h2afvb | 0.088 | actc1b | -0.049 |
| hmgb2a | 0.085 | gapdhs | -0.049 |
| mki67 | 0.084 | ptn | -0.048 |
| lbr | 0.083 | slc1a2b | -0.046 |
| ran | 0.083 | acbd7 | -0.045 |
| dut | 0.082 | glula | -0.043 |
| tuba8l4 | 0.082 | fabp7a | -0.041 |
| anp32b | 0.082 | cx43 | -0.040 |
| rrm1 | 0.082 | si:ch211-66e2.5 | -0.040 |
| cbx3a | 0.081 | ndrg3a | -0.039 |
| ccna2 | 0.080 | cotl1 | -0.039 |
| pcna | 0.080 | efhd1 | -0.039 |
| cirbpa | 0.079 | ppdpfb | -0.039 |
| seta | 0.079 | anxa13 | -0.038 |
| snrpf | 0.079 | cadm4 | -0.038 |
| snrpb | 0.078 | cox4i2 | -0.037 |
| snrpd1 | 0.077 | slc4a4a | -0.037 |
| banf1 | 0.077 | cebpd | -0.037 |
| snrpg | 0.077 | ak1 | -0.036 |
| ppm1g | 0.077 | prss59.2 | -0.036 |
| chaf1a | 0.077 | mdkb | -0.036 |
| khdrbs1a | 0.076 | cyp2ad3 | -0.036 |
| ddx39ab | 0.076 | calm1a | -0.035 |
| mcm7 | 0.076 | mt2 | -0.035 |
| snrpe | 0.075 | IGLON5 | -0.035 |
| ssrp1a | 0.075 | hbbe1.3 | -0.035 |
| dek | 0.074 | slc3a2a | -0.035 |
| rpa2 | 0.074 | si:ch211-260e23.9 | -0.035 |
| hnrnpabb | 0.073 | apoa1b | -0.035 |
| cdca5 | 0.073 | fez1 | -0.034 |
| fen1 | 0.072 | syt11a | -0.034 |
| hmgn2 | 0.072 | apoa2 | -0.034 |