ring finger protein 26
ZFIN 
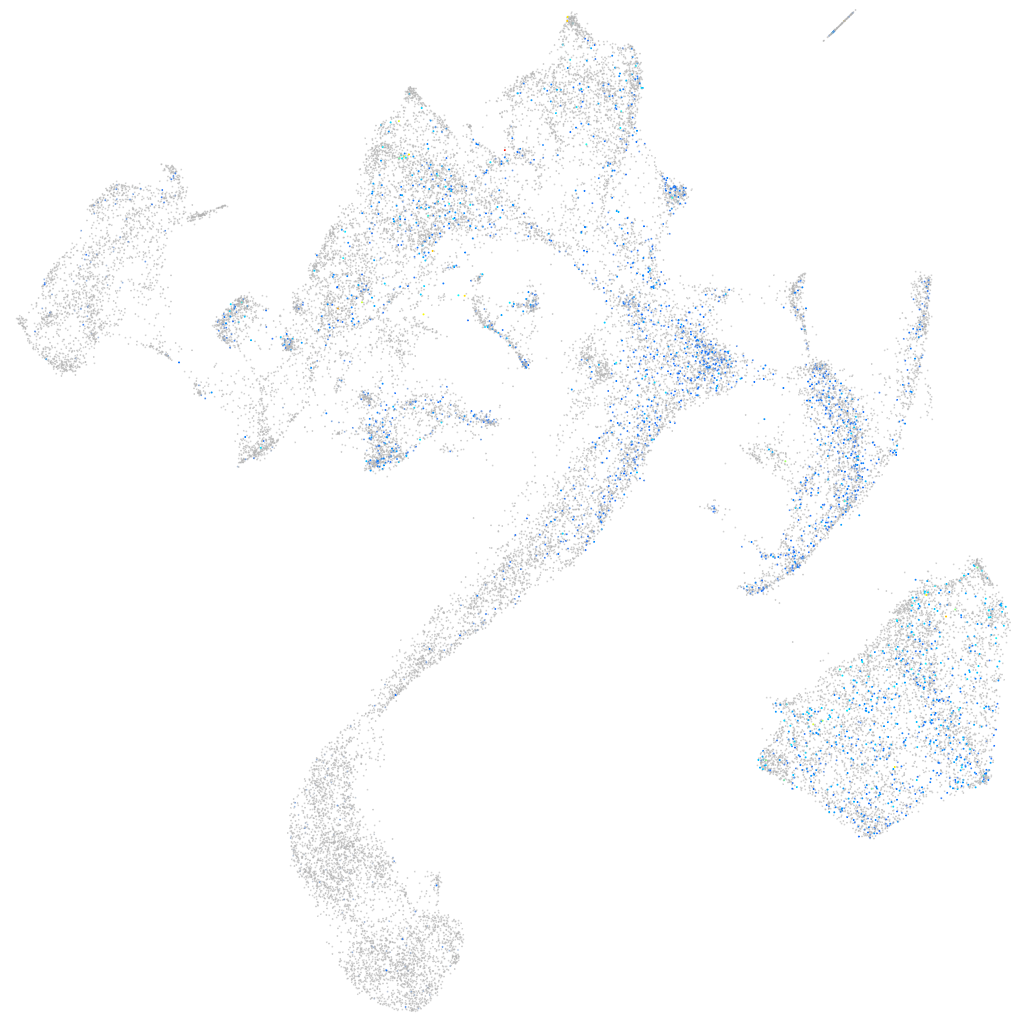
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mki67 | 0.162 | actc1b | -0.153 |
| hmgb2a | 0.159 | ckma | -0.140 |
| hmga1a | 0.156 | ckmb | -0.140 |
| si:ch73-281n10.2 | 0.152 | aldoab | -0.140 |
| ube2c | 0.152 | ak1 | -0.138 |
| anp32a | 0.151 | atp2a1 | -0.137 |
| hnrnpa0b | 0.151 | tnnc2 | -0.134 |
| si:ch211-222l21.1 | 0.151 | tmem38a | -0.132 |
| cx43.4 | 0.150 | mylpfa | -0.131 |
| cirbpa | 0.150 | neb | -0.129 |
| hnrnpaba | 0.149 | acta1b | -0.129 |
| si:ch73-1a9.3 | 0.148 | tpma | -0.126 |
| syncrip | 0.147 | ldb3b | -0.125 |
| anp32b | 0.147 | pabpc4 | -0.125 |
| hmgb2b | 0.147 | gapdh | -0.124 |
| nucks1a | 0.147 | CABZ01078594.1 | -0.123 |
| tpx2 | 0.146 | mybphb | -0.123 |
| cbx3a | 0.146 | tnnt3a | -0.122 |
| ccnb1 | 0.146 | mylz3 | -0.121 |
| aspm | 0.146 | srl | -0.121 |
| khdrbs1a | 0.146 | desma | -0.121 |
| seta | 0.145 | ttn.2 | -0.121 |
| hnrnpabb | 0.145 | eno3 | -0.120 |
| si:ch211-288g17.3 | 0.144 | actn3a | -0.120 |
| acin1a | 0.143 | ldb3a | -0.120 |
| pabpc1a | 0.142 | nme2b.2 | -0.120 |
| h2afvb | 0.141 | si:ch73-367p23.2 | -0.119 |
| ncl | 0.141 | eno1a | -0.119 |
| ptmab | 0.141 | cav3 | -0.118 |
| ctnnb1 | 0.140 | gamt | -0.118 |
| top1l | 0.140 | actn3b | -0.117 |
| nop56 | 0.139 | myl1 | -0.117 |
| plk1 | 0.139 | casq2 | -0.117 |
| ilf3b | 0.139 | ank1a | -0.116 |
| hdac1 | 0.139 | myom1a | -0.116 |