rhomboid 5 homolog 1a (Drosophila)
ZFIN 
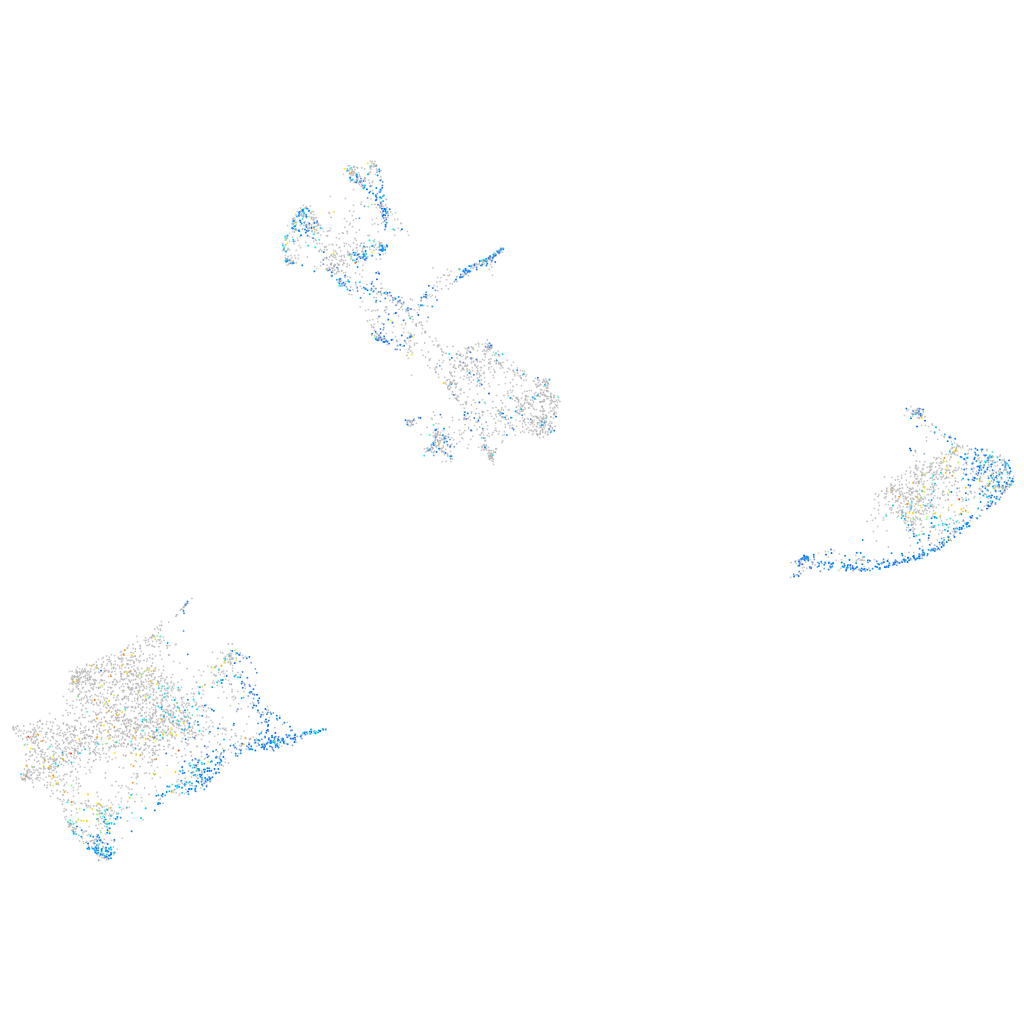
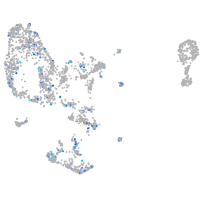
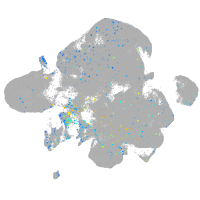
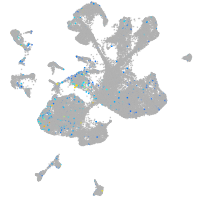
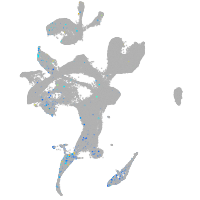
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sdc4 | 0.203 | tuba1c | -0.121 |
| trpm1b | 0.194 | ptmaa | -0.120 |
| ctsla | 0.187 | si:ch211-222l21.1 | -0.108 |
| si:zfos-943e10.1 | 0.185 | gpm6aa | -0.106 |
| gadd45bb | 0.179 | hbbe1.3 | -0.103 |
| bace2 | 0.173 | nova2 | -0.102 |
| pcdh10a | 0.170 | elavl3 | -0.102 |
| klf6a | 0.169 | stmn1b | -0.097 |
| pim1 | 0.168 | hbae3 | -0.097 |
| ctsba | 0.168 | tmsb | -0.096 |
| pttg1ipb | 0.166 | zc4h2 | -0.093 |
| sypl2b | 0.166 | gng3 | -0.093 |
| syngr1a | 0.163 | CU467822.1 | -0.091 |
| atp11a | 0.160 | hbbe1.1 | -0.091 |
| psap | 0.159 | pvalb2 | -0.090 |
| arhgef33 | 0.158 | si:ch73-1a9.3 | -0.085 |
| canx | 0.158 | fabp7a | -0.085 |
| degs1 | 0.158 | elavl4 | -0.085 |
| mitfa | 0.156 | epb41a | -0.084 |
| atp6ap1b | 0.156 | pvalb1 | -0.083 |
| ddit3 | 0.155 | zgc:65894 | -0.083 |
| BX571715.1 | 0.155 | krt4 | -0.082 |
| gpr143 | 0.155 | hmgb3a | -0.081 |
| zgc:110239 | 0.153 | sncb | -0.080 |
| id3 | 0.153 | hbae1.1 | -0.079 |
| SPAG9 | 0.152 | stx1b | -0.077 |
| inka1b | 0.152 | actc1b | -0.076 |
| tspan36 | 0.151 | cadm3 | -0.075 |
| ctsd | 0.148 | gng2 | -0.075 |
| rnaseka | 0.148 | vamp2 | -0.074 |
| opn5 | 0.147 | pbx3b | -0.071 |
| CABZ01048402.2 | 0.147 | cnrip1a | -0.071 |
| glb1l | 0.144 | hmgb1b | -0.070 |
| sptlc2a | 0.143 | mllt11 | -0.069 |
| snai1a | 0.143 | neurod1 | -0.069 |