regulator of G protein signaling 17
ZFIN 
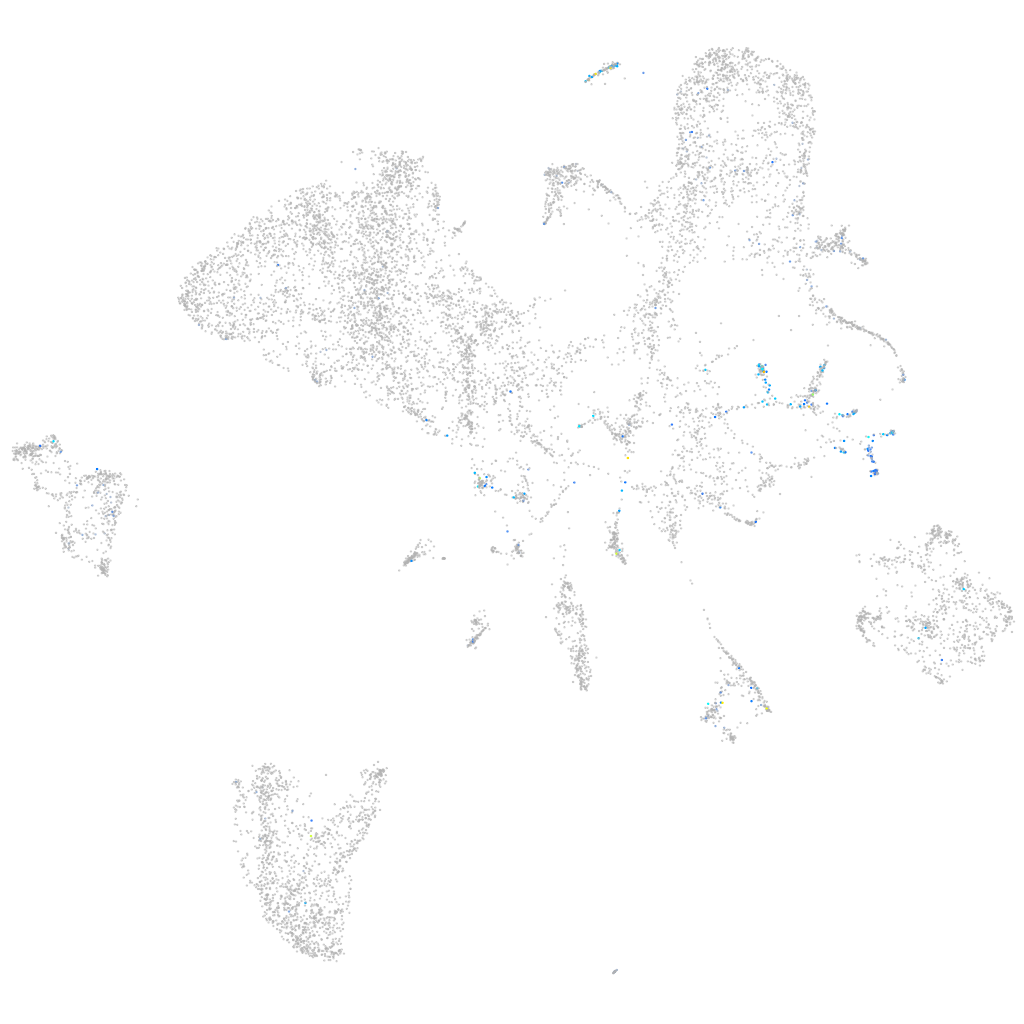
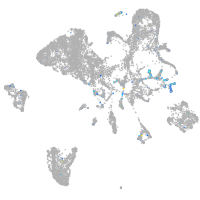
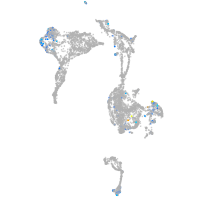
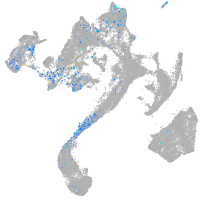
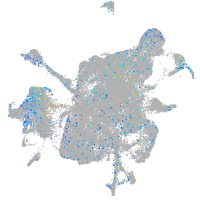
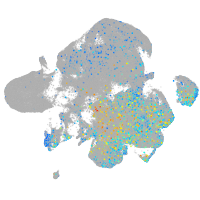
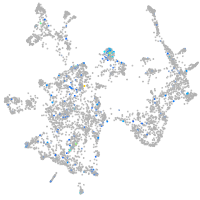
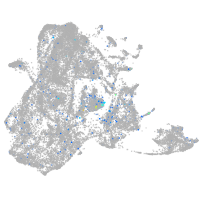
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.307 | ahcy | -0.124 |
| mir7a-1 | 0.290 | gamt | -0.121 |
| pcsk1nl | 0.289 | gapdh | -0.117 |
| pax6b | 0.284 | gatm | -0.104 |
| neurod1 | 0.258 | aldob | -0.101 |
| pcsk1 | 0.252 | bhmt | -0.098 |
| syt1a | 0.249 | fbp1b | -0.097 |
| si:dkey-153k10.9 | 0.238 | mat1a | -0.096 |
| vat1 | 0.238 | si:dkey-16p21.8 | -0.095 |
| aldocb | 0.236 | apoa1b | -0.094 |
| insm1b | 0.235 | gstt1a | -0.094 |
| fev | 0.233 | apoc2 | -0.093 |
| zgc:101731 | 0.230 | hspe1 | -0.093 |
| scgn | 0.229 | apoa4b.1 | -0.093 |
| insm1a | 0.228 | fabp3 | -0.092 |
| nkx2.2a | 0.228 | aqp12 | -0.092 |
| mir375-2 | 0.222 | gstr | -0.091 |
| lysmd2 | 0.217 | hdlbpa | -0.090 |
| rims2a | 0.216 | agxta | -0.089 |
| nrsn1 | 0.213 | afp4 | -0.088 |
| jph3 | 0.213 | cx32.3 | -0.087 |
| id4 | 0.209 | nupr1b | -0.087 |
| gpc1a | 0.208 | agxtb | -0.086 |
| spon1b | 0.206 | scp2a | -0.085 |
| hepacam2 | 0.205 | apoc1 | -0.084 |
| CABZ01072885.1 | 0.205 | aldh7a1 | -0.084 |
| gapdhs | 0.205 | pnp4b | -0.083 |
| kcnk9 | 0.205 | mgst1.2 | -0.081 |
| pygma | 0.205 | ftcd | -0.081 |
| scg2a | 0.204 | apobb.1 | -0.080 |
| slc35g2a | 0.202 | gcshb | -0.080 |
| vamp2 | 0.202 | g6pca.2 | -0.080 |
| egr4 | 0.200 | serpina1 | -0.080 |
| atp6v0cb | 0.199 | gnmt | -0.079 |
| dll4 | 0.199 | suclg2 | -0.079 |