replication factor C (activator 1) 5
ZFIN 
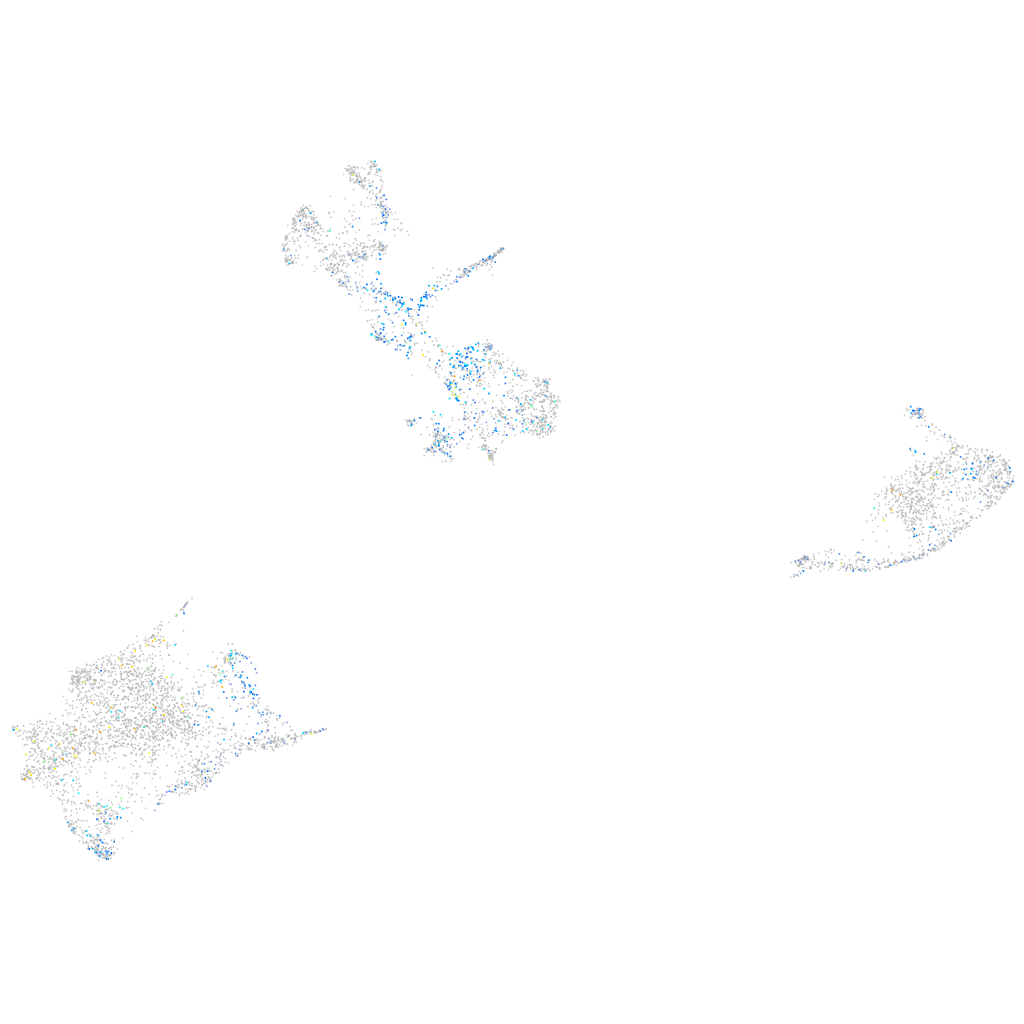
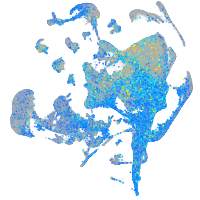
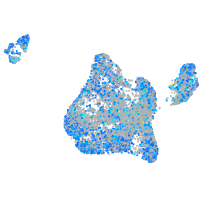
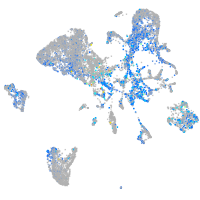
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.320 | gstp1 | -0.108 |
| dut | 0.306 | cd63 | -0.079 |
| rrm1 | 0.274 | zgc:110339 | -0.077 |
| fen1 | 0.273 | LOC103910009 | -0.077 |
| mcm7 | 0.264 | slc22a7a | -0.073 |
| nasp | 0.260 | ahnak | -0.072 |
| rpa2 | 0.257 | tspan36 | -0.071 |
| stmn1a | 0.251 | rtn1b | -0.070 |
| lig1 | 0.246 | rhag | -0.069 |
| rrm2 | 0.246 | csrp2 | -0.069 |
| si:ch211-156b7.4 | 0.240 | pah | -0.067 |
| chaf1a | 0.237 | gch2 | -0.067 |
| anp32b | 0.236 | pts | -0.064 |
| rpa3 | 0.236 | akr1b1 | -0.064 |
| mcm6 | 0.224 | si:dkey-251i10.2 | -0.064 |
| orc4 | 0.221 | XLOC-019062 | -0.063 |
| CABZ01005379.1 | 0.220 | rab38 | -0.062 |
| zgc:110540 | 0.217 | marcksl1b | -0.061 |
| cks1b | 0.214 | pvalb2 | -0.060 |
| uhrf1 | 0.214 | tspan10 | -0.060 |
| ccna2 | 0.212 | oca2 | -0.060 |
| prim2 | 0.208 | anxa1a | -0.060 |
| hmgb2b | 0.206 | kcnj13 | -0.059 |
| esco2 | 0.205 | cotl1 | -0.059 |
| dhfr | 0.204 | tyrp1b | -0.057 |
| tyms | 0.204 | slc45a2 | -0.056 |
| dek | 0.202 | mtbl | -0.056 |
| rfc4 | 0.201 | oacyl | -0.055 |
| mcm5 | 0.201 | pmela | -0.055 |
| rnaseh2a | 0.200 | myo5aa | -0.055 |
| ube2t | 0.200 | dct | -0.055 |
| cdca7a | 0.199 | slc2a11b | -0.055 |
| mcm2 | 0.198 | tyrp1a | -0.054 |
| mcm4 | 0.198 | rab27a | -0.054 |
| seta | 0.198 | zgc:91968 | -0.053 |