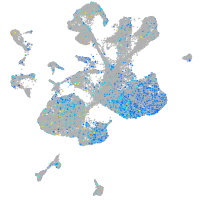
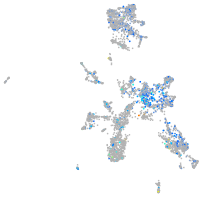
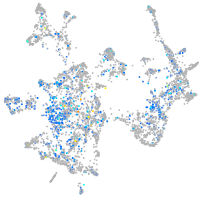
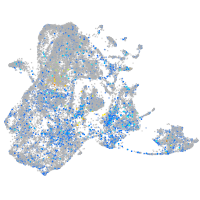
replication factor C (activator 1) 1
ZFIN 
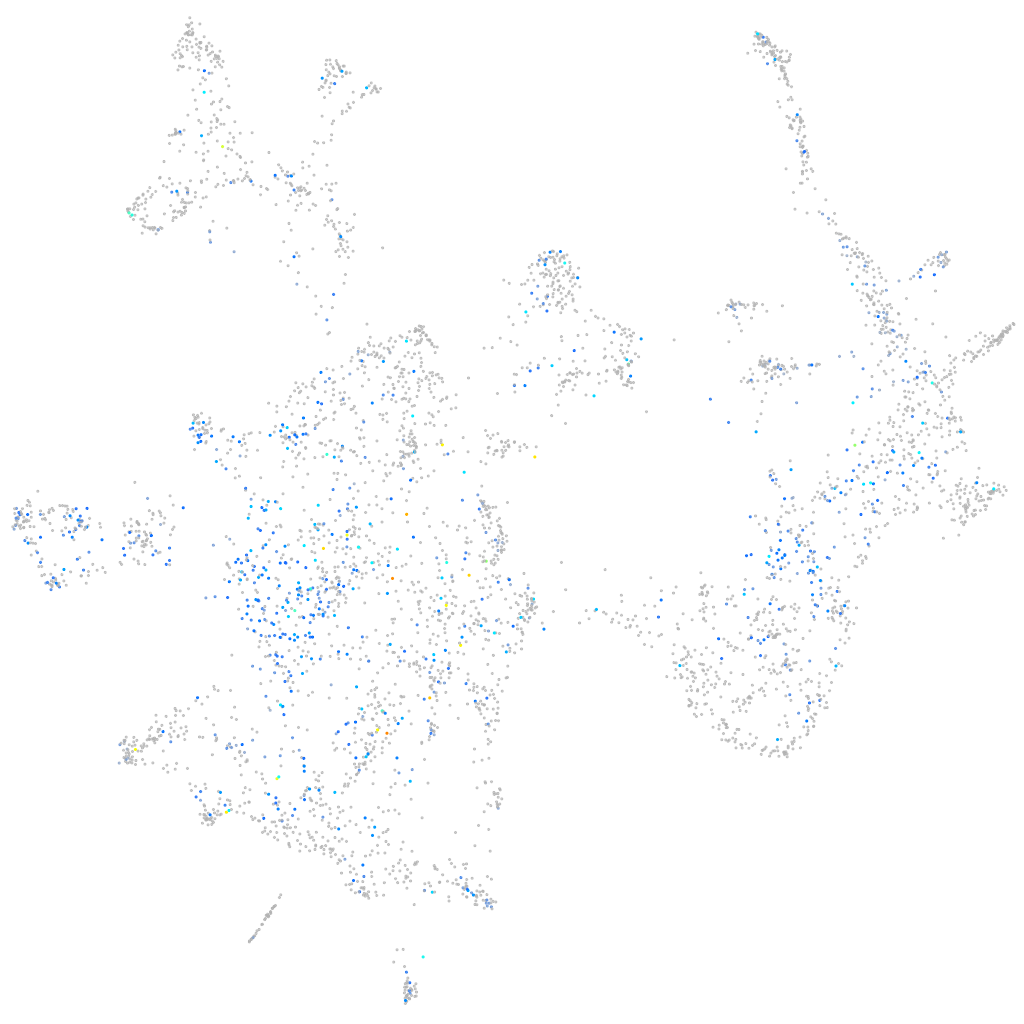
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.331 | tob1b | -0.116 |
| dut | 0.301 | h1f0 | -0.114 |
| rpa3 | 0.300 | nupr1a | -0.108 |
| lig1 | 0.295 | gabarapb | -0.100 |
| rrm1 | 0.291 | tmem59 | -0.099 |
| chaf1a | 0.286 | rnasekb | -0.097 |
| dek | 0.286 | ucp2 | -0.095 |
| rpa2 | 0.285 | pvalb8 | -0.093 |
| stmn1a | 0.282 | calm1b | -0.091 |
| fen1 | 0.281 | calm1a | -0.091 |
| zgc:110540 | 0.279 | mcl1a | -0.090 |
| nasp | 0.278 | spint2 | -0.089 |
| esco2 | 0.278 | vamp8 | -0.088 |
| CABZ01005379.1 | 0.278 | selenow1 | -0.088 |
| mcm7 | 0.273 | enosf1 | -0.087 |
| rrm2 | 0.273 | si:ch211-260e23.9 | -0.086 |
| slbp | 0.272 | pkig | -0.086 |
| rfc4 | 0.264 | capgb | -0.086 |
| si:ch211-156b7.4 | 0.260 | adipor2 | -0.086 |
| zgc:165555.3 | 0.259 | ccni | -0.086 |
| asf1ba | 0.256 | tp53inp1 | -0.085 |
| dnajc9 | 0.251 | prr15la | -0.085 |
| si:dkey-261m9.17 | 0.245 | atp6v1g1 | -0.084 |
| hist1h4l | 0.244 | smpx | -0.083 |
| anp32b | 0.243 | dedd1 | -0.082 |
| pola1 | 0.242 | atp6v1e1b | -0.082 |
| dhfr | 0.239 | eno1a | -0.082 |
| zgc:110425 | 0.239 | them6 | -0.081 |
| rbbp4 | 0.238 | hagh | -0.081 |
| mibp | 0.238 | aktip | -0.081 |
| kif15 | 0.236 | nptna | -0.081 |
| chtf18 | 0.236 | gb:bc139872 | -0.081 |
| fbxo5 | 0.235 | CR383676.1 | -0.080 |
| banf1 | 0.235 | btc | -0.080 |
| suv39h1b | 0.235 | si:dkey-280e21.3 | -0.080 |