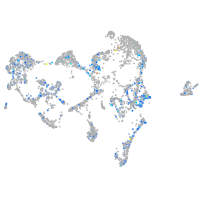
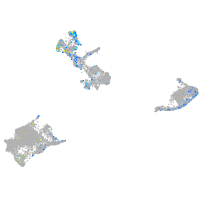
RELT like 1
ZFIN 
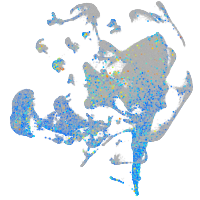
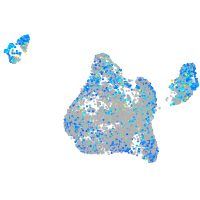
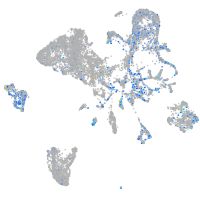
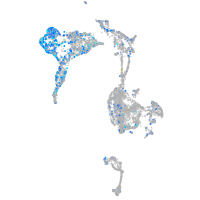
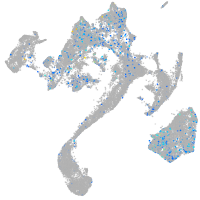
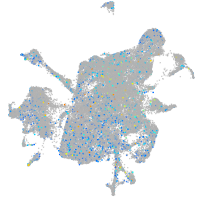
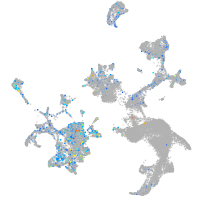
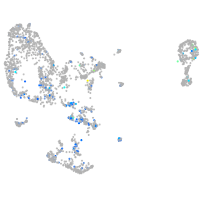
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| apoeb | 0.206 | ptmaa | -0.148 |
| apoc1 | 0.202 | rpl37 | -0.129 |
| dlx3b | 0.191 | rps10 | -0.127 |
| ved | 0.182 | tuba1c | -0.126 |
| si:ch211-152c2.3 | 0.178 | zgc:114188 | -0.124 |
| hspb1 | 0.178 | h3f3a | -0.123 |
| foxi1 | 0.175 | elavl3 | -0.115 |
| akap12b | 0.175 | hnrnpa0l | -0.112 |
| crabp2b | 0.170 | stmn1b | -0.111 |
| LOC108190024 | 0.161 | rtn1a | -0.108 |
| tagln2 | 0.161 | tmsb4x | -0.107 |
| vox | 0.160 | gpm6aa | -0.107 |
| apela | 0.160 | marcksl1a | -0.105 |
| fbl | 0.155 | fabp3 | -0.099 |
| NC-002333.4 | 0.154 | ppiab | -0.099 |
| tp63 | 0.153 | nova2 | -0.096 |
| nr6a1a | 0.152 | ckbb | -0.095 |
| npm1a | 0.151 | tmsb | -0.094 |
| nop58 | 0.149 | CR383676.1 | -0.093 |
| stm | 0.146 | si:ch1073-429i10.3.1 | -0.092 |
| dkc1 | 0.146 | gng3 | -0.085 |
| bambia | 0.146 | gpm6ab | -0.085 |
| cdh1 | 0.144 | h3f3c | -0.084 |
| tfap2c | 0.143 | rps17 | -0.084 |
| polr3gla | 0.143 | fam168a | -0.080 |
| gar1 | 0.142 | celf2 | -0.080 |
| alcamb | 0.141 | calm1a | -0.080 |
| BX927258.1 | 0.140 | tubb5 | -0.079 |
| nop2 | 0.139 | sncb | -0.078 |
| ncl | 0.139 | zgc:158463 | -0.078 |
| nop56 | 0.138 | nme2b.1 | -0.077 |
| cdx4 | 0.137 | rnasekb | -0.076 |
| lig1 | 0.136 | zc4h2 | -0.076 |
| pou5f3 | 0.135 | ppdpfb | -0.076 |
| rsl1d1 | 0.135 | ywhah | -0.076 |