RNA binding motif protein 4.3
ZFIN 
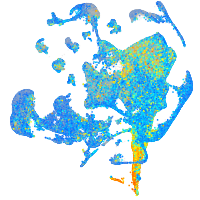
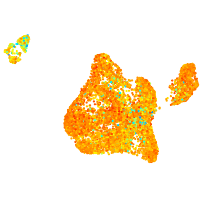
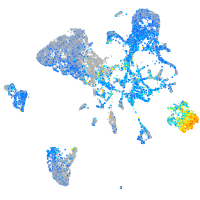
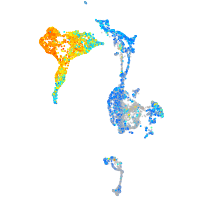
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| setb | 0.630 | rpl37 | -0.582 |
| ncl | 0.601 | ppiab | -0.578 |
| srrm1 | 0.575 | rpl8 | -0.554 |
| NC-002333.4 | 0.569 | rps10 | -0.549 |
| banf1 | 0.565 | rpl36a | -0.533 |
| stm | 0.565 | rplp2 | -0.526 |
| hnrnpa1b | 0.562 | dap1b | -0.525 |
| ccna2 | 0.560 | rps25 | -0.523 |
| syncrip | 0.556 | prdx4 | -0.517 |
| akap12b | 0.555 | rps17 | -0.513 |
| marcksb | 0.554 | rpl39 | -0.506 |
| hnrnpub | 0.550 | rplp0 | -0.502 |
| acin1b | 0.541 | prdx5 | -0.499 |
| ctsla | 0.537 | rplp1 | -0.493 |
| hnrnpaba | 0.534 | rps24 | -0.491 |
| si:ch73-281n10.2 | 0.517 | rps23 | -0.490 |
| tcea1 | 0.510 | zgc:114188 | -0.489 |
| snrpb2 | 0.506 | rps5 | -0.488 |
| sf3b2 | 0.505 | eef1db | -0.488 |
| mki67 | 0.505 | NDUFB1 | -0.486 |
| kmt5ab | 0.487 | rpl22 | -0.485 |
| gar1 | 0.487 | mtx2 | -0.481 |
| h1m | 0.486 | rps14 | -0.481 |
| marcksl1b | 0.482 | rpl11 | -0.477 |
| zmat2 | 0.480 | cox6a1 | -0.476 |
| snrpa | 0.480 | rpl21 | -0.475 |
| cx43.4 | 0.479 | rpl13a | -0.474 |
| nop56 | 0.478 | rps27.1 | -0.468 |
| ppig | 0.473 | rps16 | -0.468 |
| hnrnpabb | 0.472 | faua | -0.462 |
| anp32a | 0.466 | rps9 | -0.461 |
| ddit4 | 0.463 | cd63 | -0.458 |
| tbx16 | 0.456 | ISCU (1 of many) | -0.457 |
| hnrnpl | 0.455 | rpl9 | -0.454 |
| sart3 | 0.453 | rnasekb | -0.452 |