ribokinase
ZFIN 
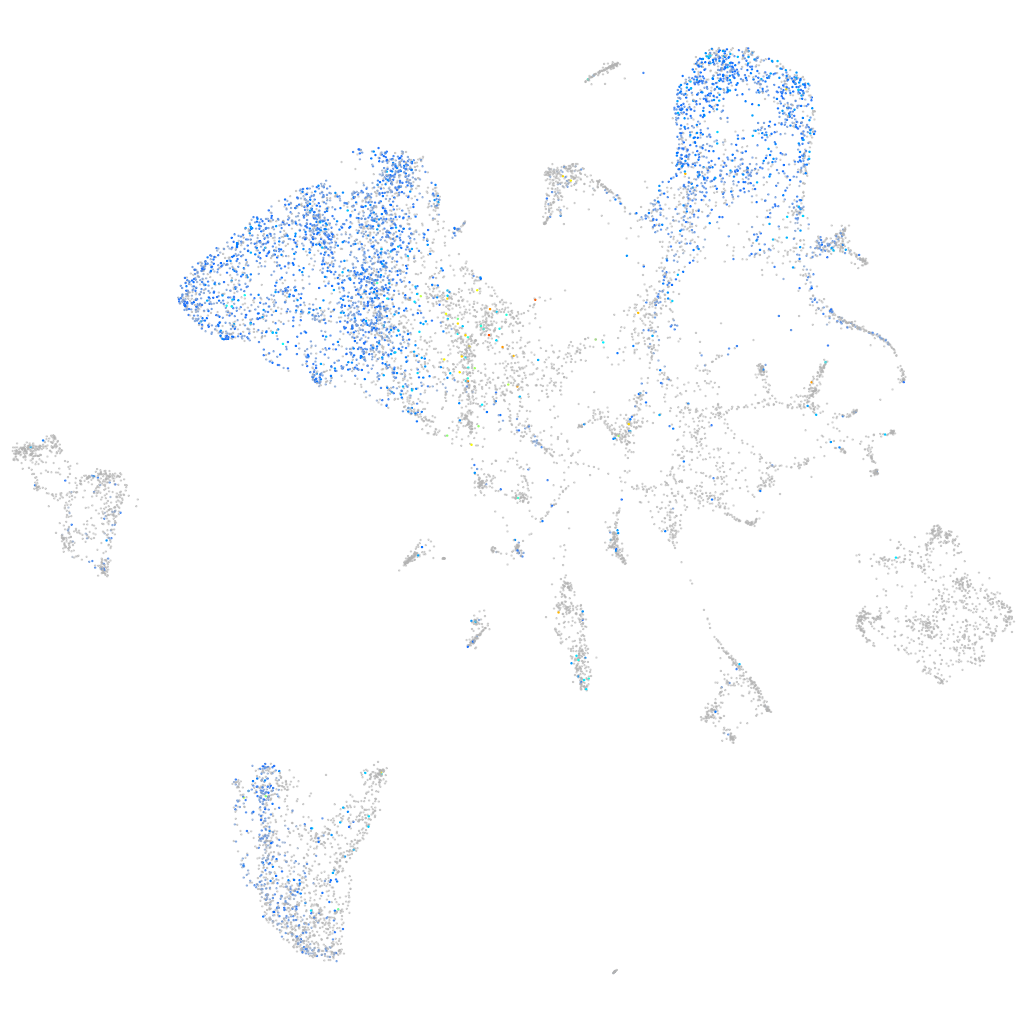
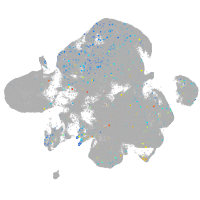
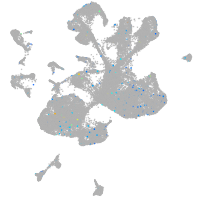
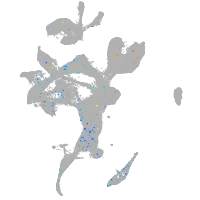
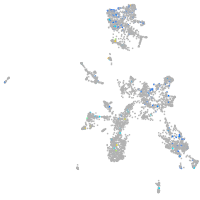
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| glud1b | 0.422 | hmgb3a | -0.227 |
| rdh1 | 0.404 | hmgn6 | -0.226 |
| scp2a | 0.400 | si:ch211-222l21.1 | -0.225 |
| apoa4b.1 | 0.397 | epcam | -0.221 |
| dhrs9 | 0.394 | marcksl1b | -0.219 |
| sod1 | 0.390 | jpt1b | -0.219 |
| srd5a2a | 0.389 | h3f3d | -0.216 |
| gpx4a | 0.389 | ywhaz | -0.207 |
| gapdh | 0.389 | si:ch73-1a9.3 | -0.204 |
| fbp1b | 0.386 | cldnb | -0.202 |
| cyp8b1 | 0.385 | glulb | -0.201 |
| sult2st2 | 0.384 | marcksb | -0.199 |
| apoc2 | 0.383 | hmgb1b | -0.196 |
| suclg1 | 0.380 | cldn7b | -0.193 |
| slc37a4a | 0.380 | ubc | -0.193 |
| mdh1aa | 0.378 | nucks1a | -0.191 |
| gcshb | 0.377 | id1 | -0.190 |
| ugt1a7 | 0.376 | hmgb1a | -0.188 |
| ugt5b4 | 0.373 | hmgb2a | -0.188 |
| eno3 | 0.370 | hnrnpaba | -0.188 |
| gstt1a | 0.364 | syncrip | -0.186 |
| mgst1.2 | 0.363 | si:ch73-281n10.2 | -0.185 |
| fdx1 | 0.363 | aldoaa | -0.185 |
| apobb.1 | 0.359 | bcam | -0.185 |
| gstr | 0.356 | khdrbs1a | -0.184 |
| haao | 0.356 | ptmab | -0.184 |
| acaa1 | 0.355 | cirbpb | -0.182 |
| cyp4v8 | 0.354 | h2afvb | -0.182 |
| sult1st6 | 0.354 | hnrnpa0a | -0.181 |
| sdr16c5b | 0.352 | tuba8l4 | -0.181 |
| pgam1a | 0.351 | hmgn2 | -0.180 |
| acadm | 0.351 | btg1 | -0.180 |
| abat | 0.350 | cx43.4 | -0.180 |
| sod2 | 0.349 | acin1a | -0.179 |
| cx32.3 | 0.348 | cd9b | -0.178 |