RNA binding fox-1 homolog 3b
ZFIN 
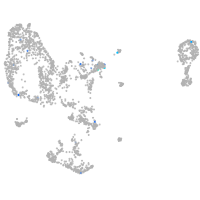
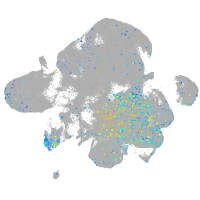
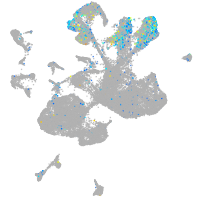
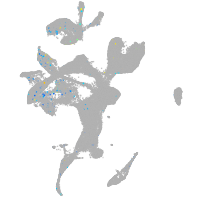
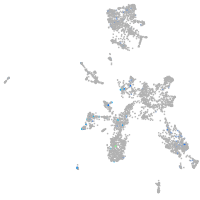
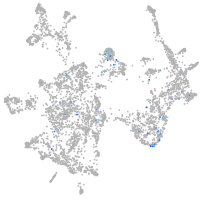
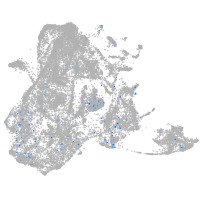
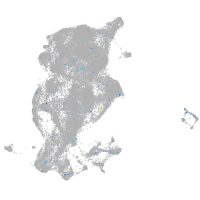
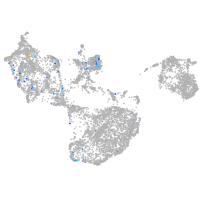
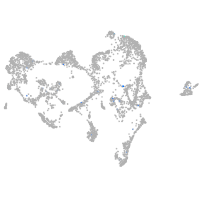
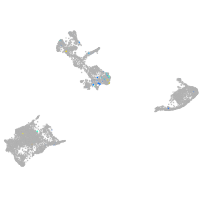
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mnx1 | 0.795 | pnrc2 | -0.265 |
| nkx6.2 | 0.795 | mt-co1 | -0.237 |
| LOC103910736 | 0.795 | rtn4a | -0.215 |
| insm1a | 0.791 | kpna4 | -0.215 |
| ins | 0.783 | rbm8a | -0.215 |
| col9a1b | 0.780 | syncrip | -0.215 |
| LOC101883836 | 0.760 | sde2 | -0.211 |
| atcaya | 0.745 | rtca | -0.200 |
| rab7b | 0.744 | rps18 | -0.198 |
| CT573204.1 | 0.739 | hnrnph1l | -0.197 |
| myl9a | 0.738 | ncl | -0.193 |
| map6a | 0.736 | stmn1a | -0.193 |
| rsph9 | 0.736 | ptges3b | -0.192 |
| gng12a | 0.731 | hmgb2a | -0.188 |
| pax6b | 0.700 | nkap | -0.187 |
| si:ch73-25f10.6 | 0.683 | seta | -0.187 |
| slc26a6 | 0.675 | sltm | -0.186 |
| mllt11 | 0.662 | bzw1b | -0.184 |
| myl10 | 0.655 | ctsla | -0.184 |
| arfip2a | 0.641 | msna | -0.183 |
| sgpl1 | 0.618 | si:ch73-281n10.2 | -0.183 |
| parvb | 0.611 | anp32b | -0.182 |
| ece1 | 0.605 | zgc:56699 | -0.182 |
| neurod1 | 0.599 | ranbp1 | -0.180 |
| thrap3a | 0.597 | h2afva | -0.180 |
| nudt9 | 0.596 | snrnp70 | -0.180 |
| itgb1b.1 | 0.587 | srrm1 | -0.180 |
| st3gal7 | 0.549 | ywhae1 | -0.179 |
| gng2 | 0.543 | setb | -0.179 |
| mtss1la | 0.543 | nelfe | -0.178 |
| greb1l | 0.542 | dek | -0.172 |
| LOC101883391 | 0.542 | sap30bp | -0.172 |
| ntd5 | 0.538 | zmat2 | -0.170 |
| si:dkey-11f4.14 | 0.536 | spcs2 | -0.170 |
| ankrd46a | 0.533 | ppiaa | -0.170 |