Ras association domain family member 8a
ZFIN 
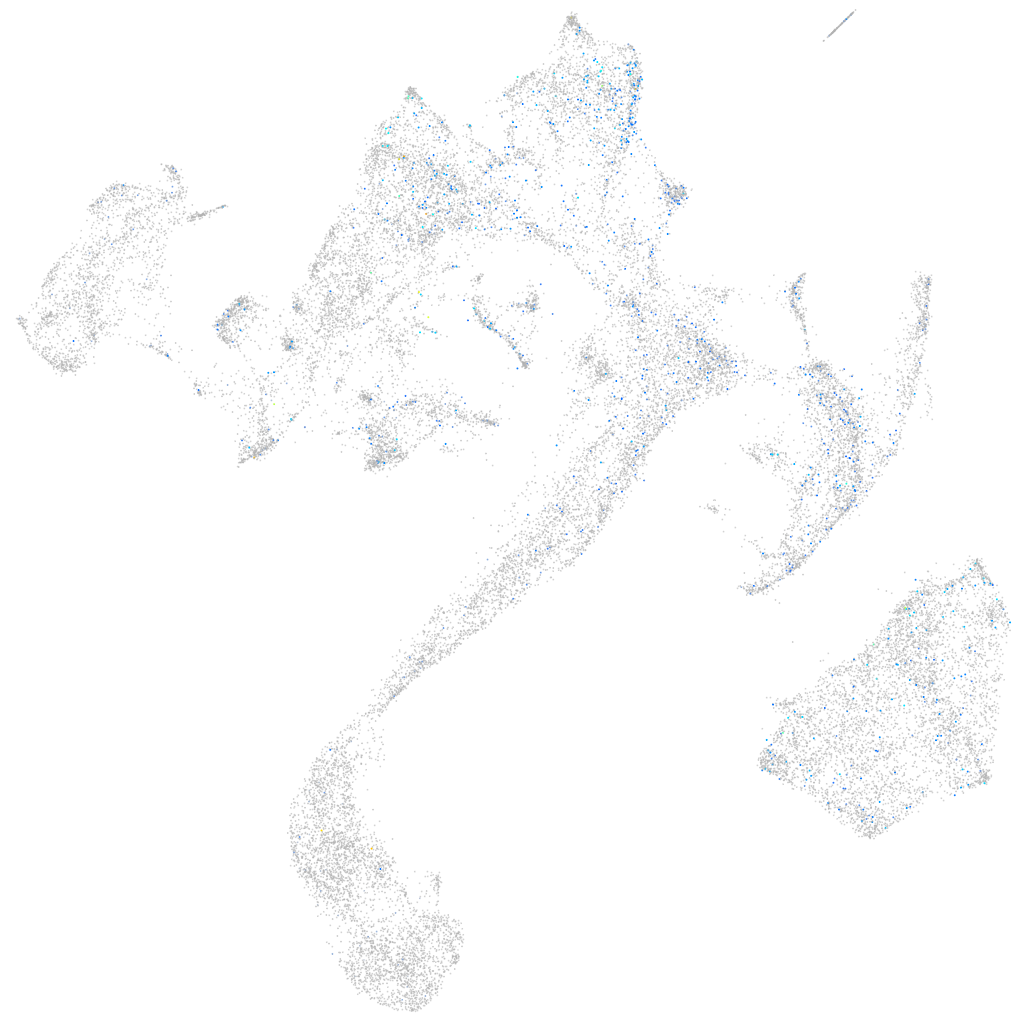
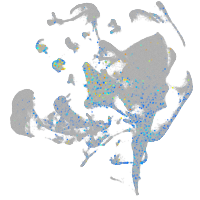
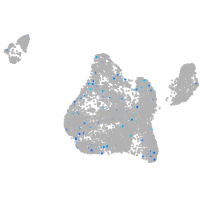
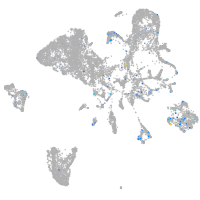
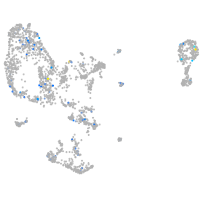
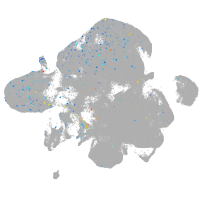
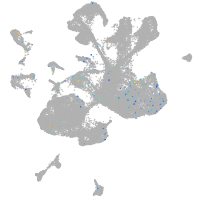
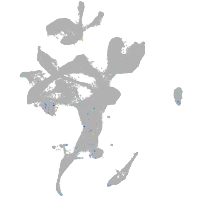
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ppib | 0.096 | actc1b | -0.082 |
| colec12 | 0.093 | aldoab | -0.079 |
| tpm4a | 0.093 | ak1 | -0.077 |
| fkbp7 | 0.092 | tmem38a | -0.077 |
| col11a1b | 0.092 | ckma | -0.076 |
| col5a1 | 0.091 | atp2a1 | -0.076 |
| marcksl1a | 0.091 | tnnc2 | -0.076 |
| col1a1b | 0.089 | pabpc4 | -0.075 |
| fkbp9 | 0.089 | ckmb | -0.075 |
| mfap2 | 0.088 | gapdh | -0.073 |
| si:rp71-17i16.5 | 0.087 | acta1b | -0.073 |
| col5a2a | 0.087 | tpma | -0.072 |
| reck | 0.086 | ttn.2 | -0.072 |
| actb2 | 0.085 | mybphb | -0.072 |
| pmp22a | 0.085 | srl | -0.071 |
| col1a2 | 0.085 | acta1a | -0.071 |
| gulp1a | 0.084 | atp5if1b | -0.070 |
| actb1 | 0.084 | ldb3b | -0.070 |
| fkbp14 | 0.083 | neb | -0.070 |
| nkx3.2 | 0.083 | CABZ01078594.1 | -0.070 |
| tgfbi | 0.082 | desma | -0.069 |
| pax9 | 0.082 | ttn.1 | -0.069 |
| CU929237.1 | 0.081 | hhatla | -0.069 |
| ssr3 | 0.081 | myl1 | -0.068 |
| col1a1a | 0.080 | smyd1a | -0.068 |
| CR749762.1 | 0.080 | actn3a | -0.068 |
| cthrc1a | 0.080 | cav3 | -0.067 |
| col2a1b | 0.080 | gatm | -0.067 |
| foxc1b | 0.079 | eno1a | -0.067 |
| tmsb4x | 0.079 | cox17 | -0.067 |
| cd82a | 0.079 | mylpfa | -0.067 |
| mmp2 | 0.079 | cycsb | -0.066 |
| krt18a.1 | 0.079 | klhl41b | -0.066 |
| ssr2 | 0.078 | gamt | -0.066 |
| hmgn2 | 0.078 | ank1a | -0.066 |