RAS guanyl releasing protein 3 (calcium and DAG-regulated)
ZFIN 
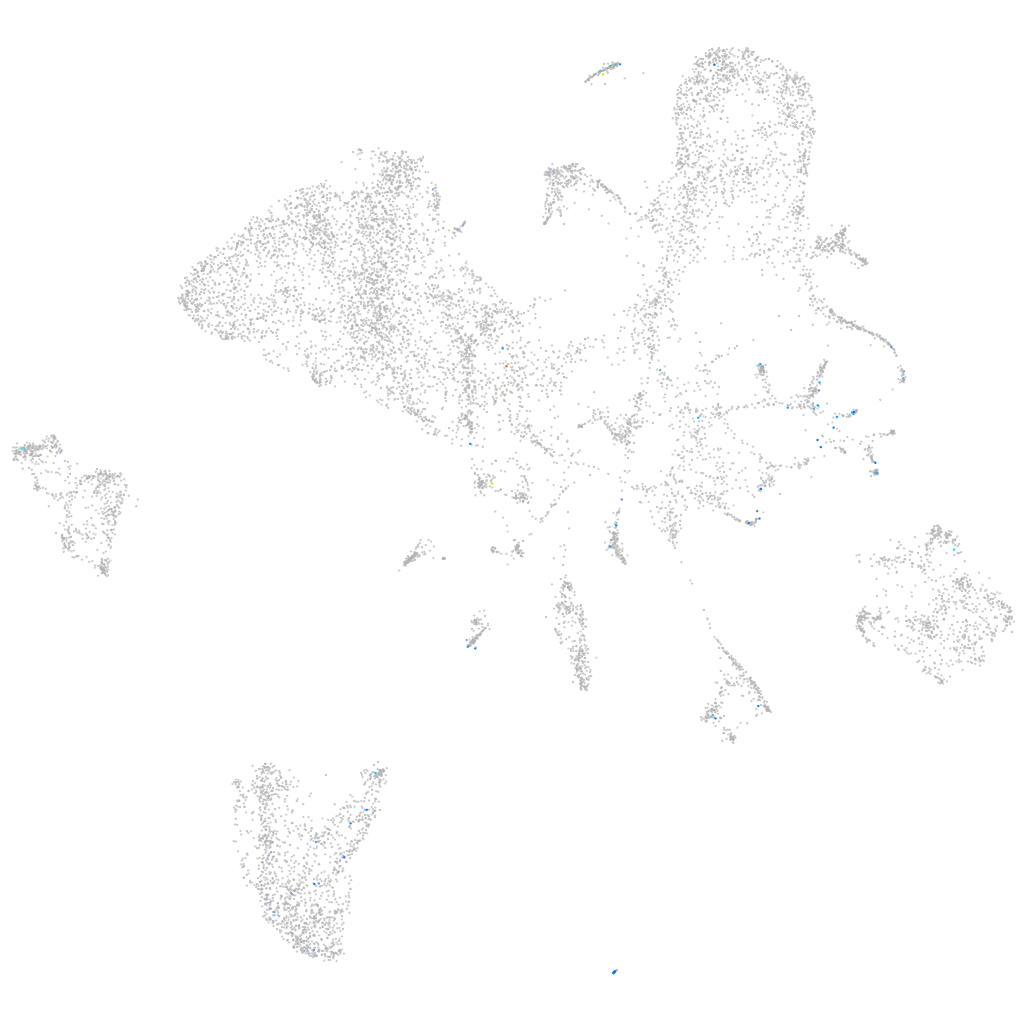
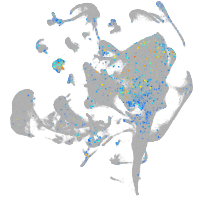
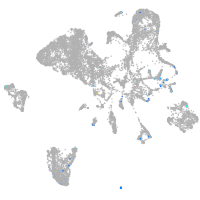
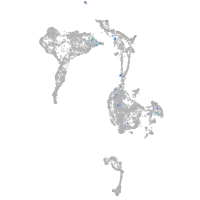
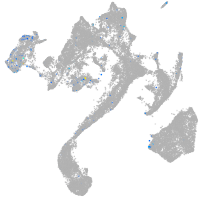
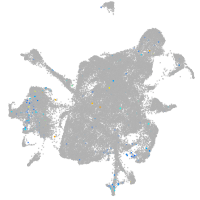
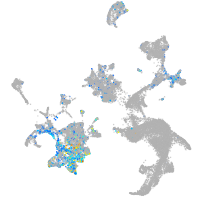
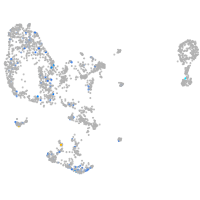
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kcnj10a | 0.237 | aldob | -0.069 |
| XLOC-020238 | 0.203 | apoc2 | -0.066 |
| CT025771.1 | 0.200 | apoa4b.1 | -0.065 |
| LOC101883142 | 0.197 | gstt1a | -0.062 |
| hyal6 | 0.189 | gapdh | -0.060 |
| galr1a | 0.186 | gstr | -0.059 |
| LOC100151570 | 0.177 | afp4 | -0.059 |
| ca14 | 0.171 | haao | -0.058 |
| CU570682.1 | 0.170 | apoa1b | -0.056 |
| ccl38.1 | 0.169 | hspd1 | -0.056 |
| tg | 0.159 | apoc1 | -0.055 |
| gas2a | 0.157 | ugt1a7 | -0.054 |
| tph2 | 0.150 | apobb.1 | -0.054 |
| slc5a5 | 0.148 | fbp1b | -0.054 |
| si:ch211-165i18.2 | 0.144 | sdr16c5b | -0.054 |
| slco1c1 | 0.141 | ahcy | -0.054 |
| pparda | 0.141 | tdo2a | -0.054 |
| tagapb | 0.140 | suclg1 | -0.053 |
| nkx2.4b | 0.140 | scp2a | -0.052 |
| tpo | 0.137 | suclg2 | -0.052 |
| MAPK8IP1 (1 of many) | 0.137 | ambp | -0.052 |
| mlc1 | 0.137 | apom | -0.052 |
| penka | 0.134 | igfbp2a | -0.052 |
| nmbb | 0.132 | eno3 | -0.052 |
| duox2 | 0.132 | lipf | -0.052 |
| anks1ab | 0.131 | si:dkey-16p21.8 | -0.052 |
| phex | 0.130 | gamt | -0.052 |
| slc8a2a | 0.130 | aldh9a1a.1 | -0.051 |
| si:dkey-42p14.3 | 0.129 | msrb2 | -0.051 |
| si:dkey-7n6.2 | 0.127 | prdx2 | -0.051 |
| moxd1l | 0.125 | rplp2l | -0.051 |
| SLC46A3 (1 of many) | 0.124 | upb1 | -0.051 |
| si:dkey-211g8.6 | 0.124 | acaa2 | -0.051 |
| soga3b | 0.123 | si:ch1073-325m22.2 | -0.051 |
| ccdc3a | 0.123 | ftcd | -0.051 |