RASD family member 3
ZFIN 
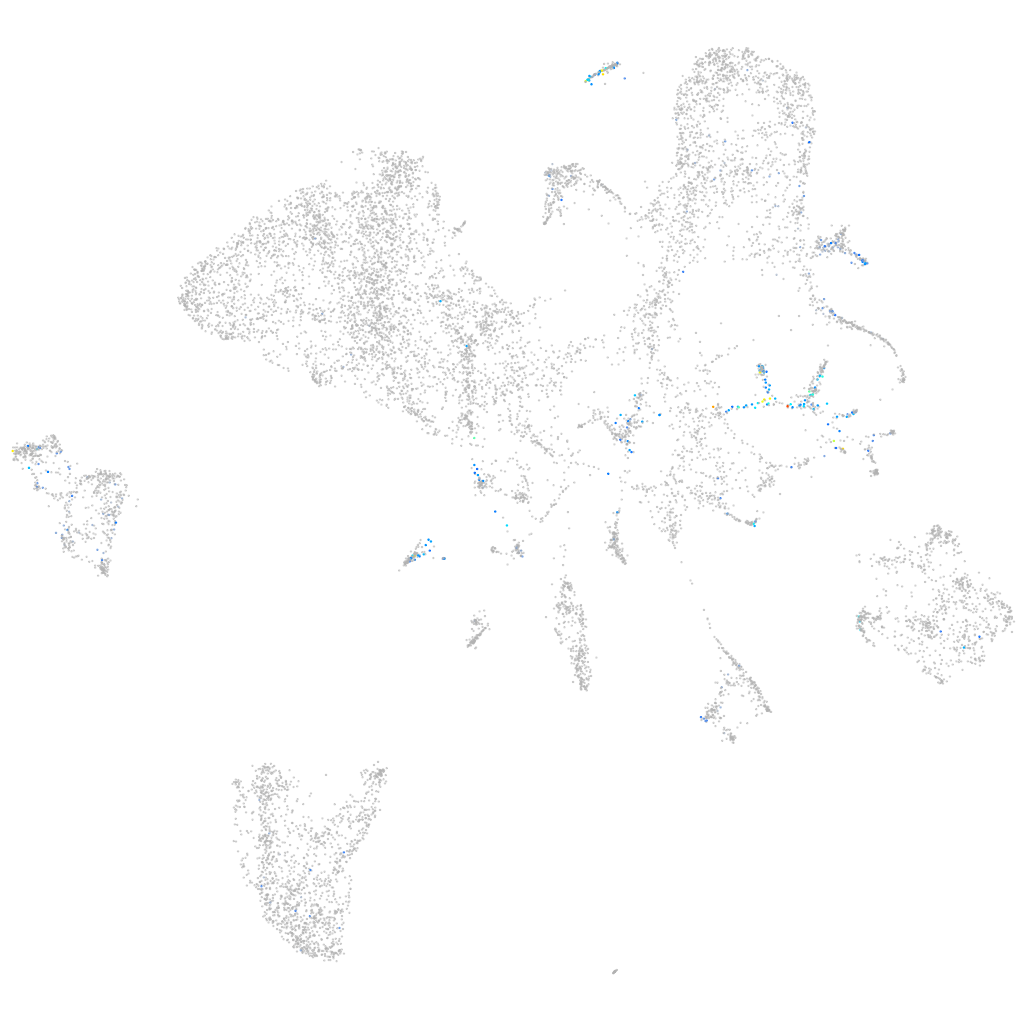
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fev | 0.316 | gamt | -0.130 |
| insm1b | 0.300 | gapdh | -0.117 |
| dll4 | 0.287 | gatm | -0.112 |
| pax4 | 0.279 | ahcy | -0.107 |
| neurod1 | 0.279 | fbp1b | -0.107 |
| hepacam2 | 0.279 | bhmt | -0.107 |
| scg3 | 0.272 | apoa1b | -0.103 |
| mir7a-1 | 0.270 | apoc2 | -0.103 |
| zgc:101731 | 0.269 | mat1a | -0.102 |
| mir375-2 | 0.263 | apoa4b.1 | -0.101 |
| neurog3 | 0.251 | aldob | -0.100 |
| gfra3 | 0.247 | afp4 | -0.099 |
| egr4 | 0.238 | aldh7a1 | -0.098 |
| elovl1a | 0.235 | aqp12 | -0.098 |
| dlb | 0.223 | gstt1a | -0.095 |
| hpcal1 | 0.220 | scp2a | -0.094 |
| fhl1b | 0.216 | apoc1 | -0.093 |
| XLOC-007078 | 0.215 | apoa2 | -0.093 |
| tmsb | 0.212 | aldh6a1 | -0.092 |
| slc7a14a | 0.208 | cx32.3 | -0.092 |
| anxa4 | 0.203 | cdo1 | -0.092 |
| dusp2 | 0.200 | agxtb | -0.091 |
| vamp2 | 0.200 | si:dkey-16p21.8 | -0.091 |
| insm1a | 0.197 | apobb.1 | -0.090 |
| adcyap1a | 0.196 | mgst1.2 | -0.089 |
| id4 | 0.196 | hdlbpa | -0.087 |
| pcsk1nl | 0.194 | agxta | -0.087 |
| syt1a | 0.193 | nupr1b | -0.087 |
| gdi1 | 0.193 | wu:fj16a03 | -0.087 |
| lmx1ba | 0.193 | slc27a2a | -0.087 |
| nkx2.2a | 0.193 | hspd1 | -0.086 |
| pygma | 0.190 | gcshb | -0.086 |
| cldnh | 0.189 | gstr | -0.085 |
| rgs16 | 0.187 | fabp3 | -0.085 |
| LOC110438375 | 0.186 | tfa | -0.085 |