Ral GEF with PH domain and SH3 binding motif 2
ZFIN 
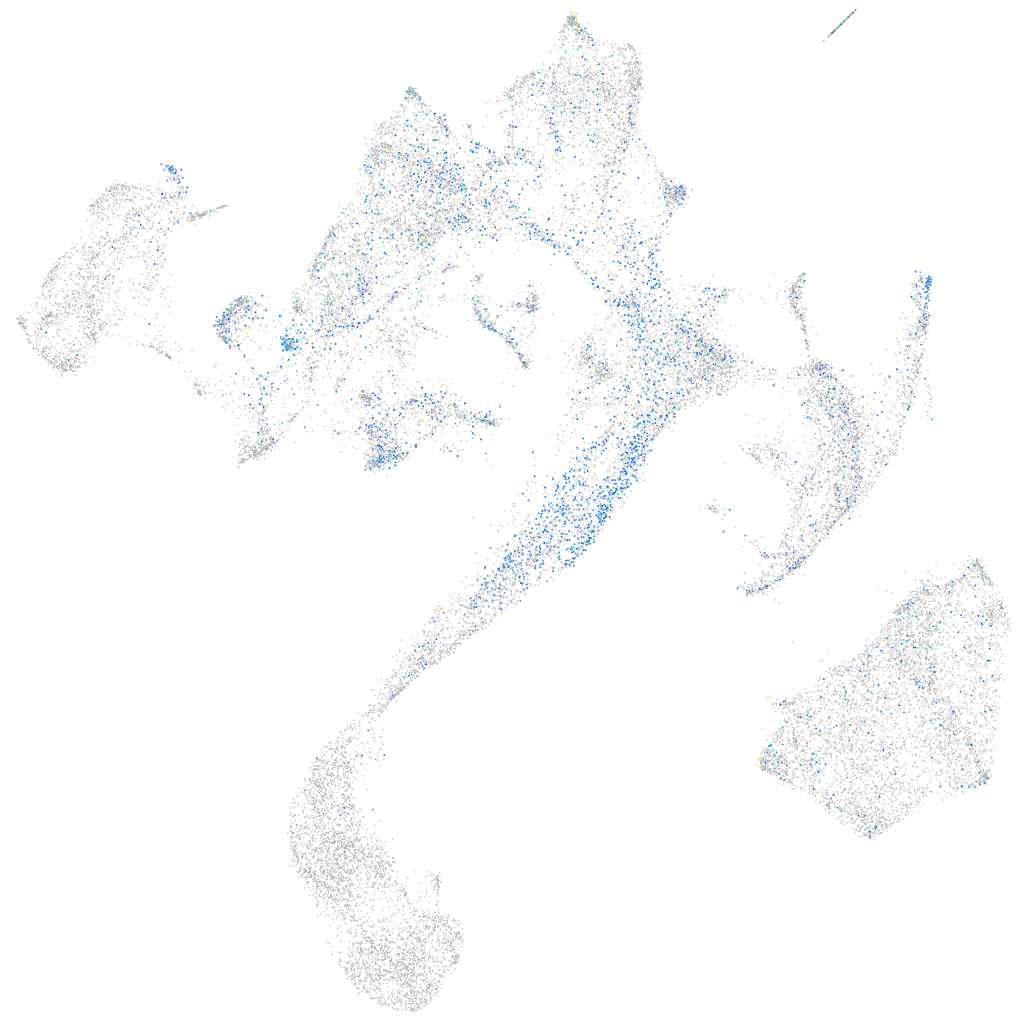
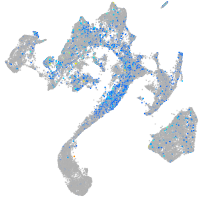
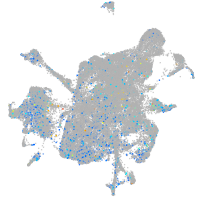
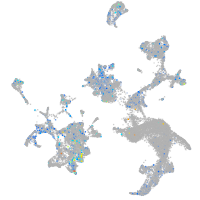
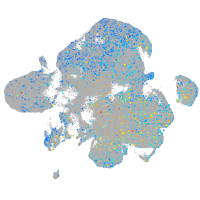
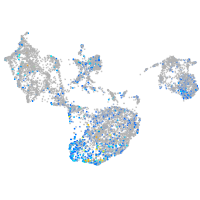
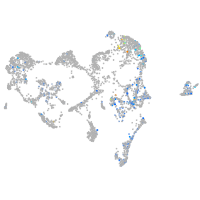
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| jam2a | 0.165 | myom1a | -0.116 |
| mafbb | 0.161 | smyd1a | -0.116 |
| si:ch211-43f4.1 | 0.161 | si:ch73-367p23.2 | -0.115 |
| myog | 0.159 | actn3a | -0.115 |
| fam107b | 0.159 | pgam2 | -0.114 |
| rbm24a | 0.150 | si:ch211-255p10.3 | -0.112 |
| zbtb18 | 0.145 | si:dkey-16p21.8 | -0.112 |
| tcf12 | 0.142 | mylpfb | -0.110 |
| cd81a | 0.141 | pkmb | -0.110 |
| si:ch1073-268j14.1 | 0.141 | gapdh | -0.110 |
| eef2b | 0.139 | pvalb1 | -0.110 |
| vwde | 0.139 | tmod4 | -0.109 |
| midn | 0.138 | XLOC-025819 | -0.109 |
| emp2 | 0.138 | ckmb | -0.109 |
| lmnb1 | 0.137 | prx | -0.109 |
| hspa8 | 0.135 | ckma | -0.109 |
| thrap3a | 0.134 | ank1a | -0.108 |
| jpt1b | 0.133 | XLOC-001975 | -0.107 |
| cxxc5a | 0.133 | tnni2a.4 | -0.107 |
| megf10 | 0.132 | dhrs7cb | -0.107 |
| pkig | 0.131 | XLOC-006515 | -0.107 |
| rps9 | 0.131 | pvalb2 | -0.106 |
| zgc:56493 | 0.130 | mylz3 | -0.105 |
| mdka | 0.130 | casq1b | -0.105 |
| cygb1 | 0.129 | XLOC-005350 | -0.105 |
| her6 | 0.129 | eef2l2 | -0.104 |
| fam49a | 0.128 | myoz1b | -0.104 |
| rpl12 | 0.128 | si:ch211-266g18.10 | -0.104 |
| rps14 | 0.128 | slc25a4 | -0.103 |
| aktip | 0.127 | myom2a | -0.102 |
| igfbp5b | 0.127 | CABZ01061524.1 | -0.102 |
| hnrnpa0l | 0.127 | eno3 | -0.102 |
| rpl13 | 0.126 | ldb3b | -0.101 |
| tenm3 | 0.126 | cavin4a | -0.101 |
| mef2d | 0.125 | neb | -0.101 |