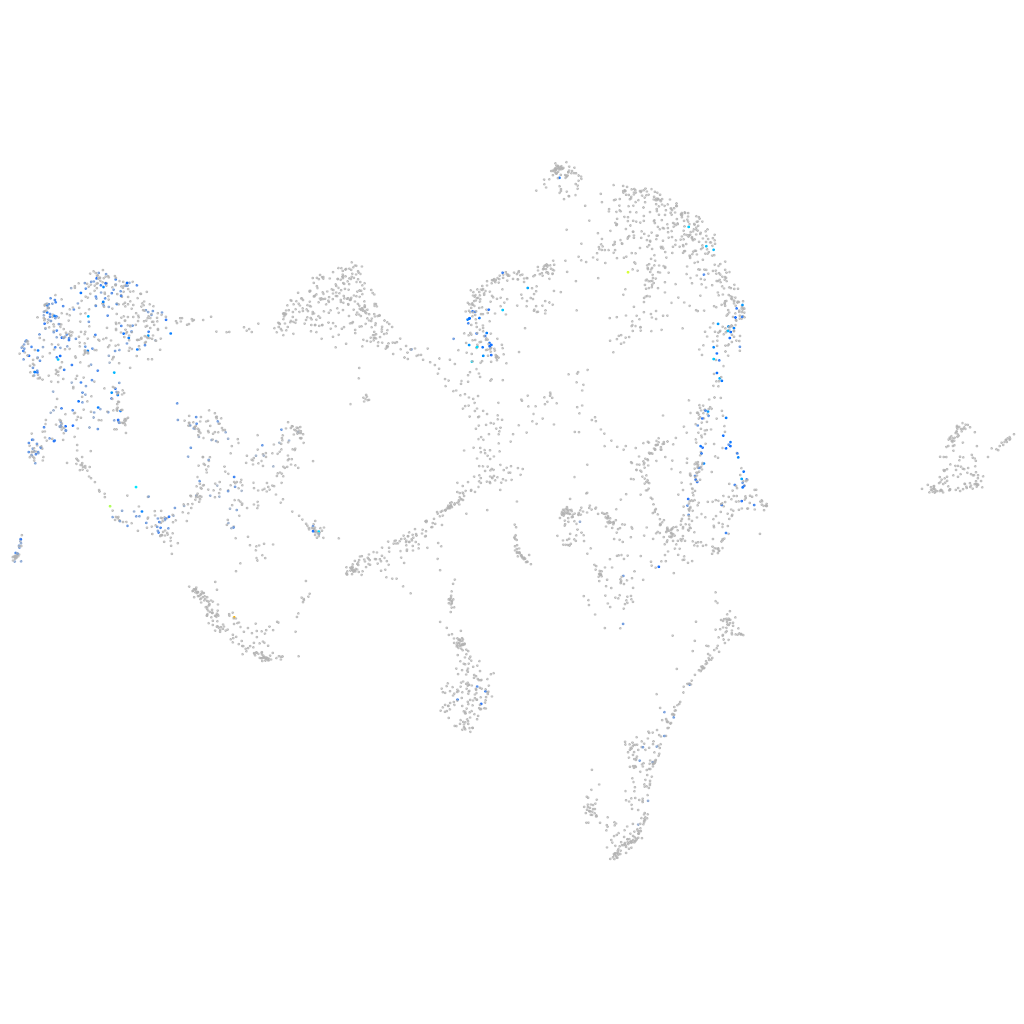
RAB GTPase activating protein 1-like 2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm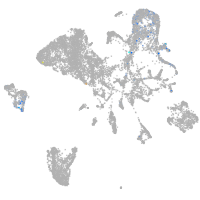 |
axial mesoderm |
muscle 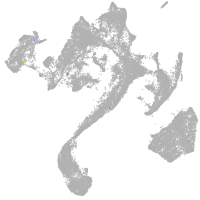 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin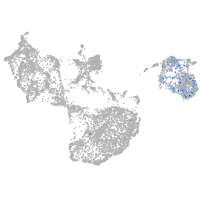 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:163030 | 0.249 | id3 | -0.126 |
| lye | 0.247 | cotl1 | -0.108 |
| fut9d | 0.246 | COX3 | -0.098 |
| dhrs13a.2 | 0.245 | ppiaa | -0.094 |
| s100w | 0.243 | dbi | -0.093 |
| ckap4 | 0.242 | vdac3 | -0.093 |
| si:ch73-52f15.5 | 0.240 | mt-atp6 | -0.092 |
| cyt1l | 0.239 | gcm2 | -0.092 |
| tcnbb | 0.237 | ndufs2 | -0.091 |
| zgc:193505 | 0.235 | ndufb5 | -0.091 |
| si:ch211-157c3.4 | 0.234 | rac2 | -0.091 |
| si:ch211-217k17.9 | 0.234 | mt-co2 | -0.090 |
| FO904880.1 | 0.232 | fthl27 | -0.090 |
| si:dkey-222n6.2 | 0.231 | pax9 | -0.089 |
| cyt1 | 0.230 | txndc9 | -0.088 |
| agr1 | 0.229 | sparc | -0.088 |
| dhrs11a | 0.229 | degs1 | -0.086 |
| anxa1c | 0.228 | atp5fa1 | -0.085 |
| FP236513.1 | 0.225 | ppifb | -0.084 |
| krt4 | 0.223 | mt-nd1 | -0.084 |
| anxa1b | 0.222 | sdha | -0.084 |
| stx11b.1 | 0.221 | kdm6bb | -0.083 |
| si:ch211-125o16.4 | 0.219 | foxi3a | -0.083 |
| cx30.3 | 0.217 | sat1a.2 | -0.083 |
| tcima | 0.215 | atp6v1aa | -0.082 |
| wu:fb18f06 | 0.208 | zgc:158463 | -0.082 |
| mid1ip1a | 0.207 | atp5mf | -0.081 |
| zgc:175088 | 0.207 | cox7a2a | -0.080 |
| si:ch211-195b11.3 | 0.206 | got2b | -0.080 |
| icn2 | 0.203 | mrpl48 | -0.080 |
| CABZ01099847.1 | 0.200 | aldh7a1 | -0.080 |
| pnp5a | 0.199 | atp5if1a | -0.080 |
| BX901957.1 | 0.198 | gapdhs | -0.079 |
| evpla | 0.197 | cldni | -0.079 |
| zgc:174938 | 0.194 | hsd17b10 | -0.079 |