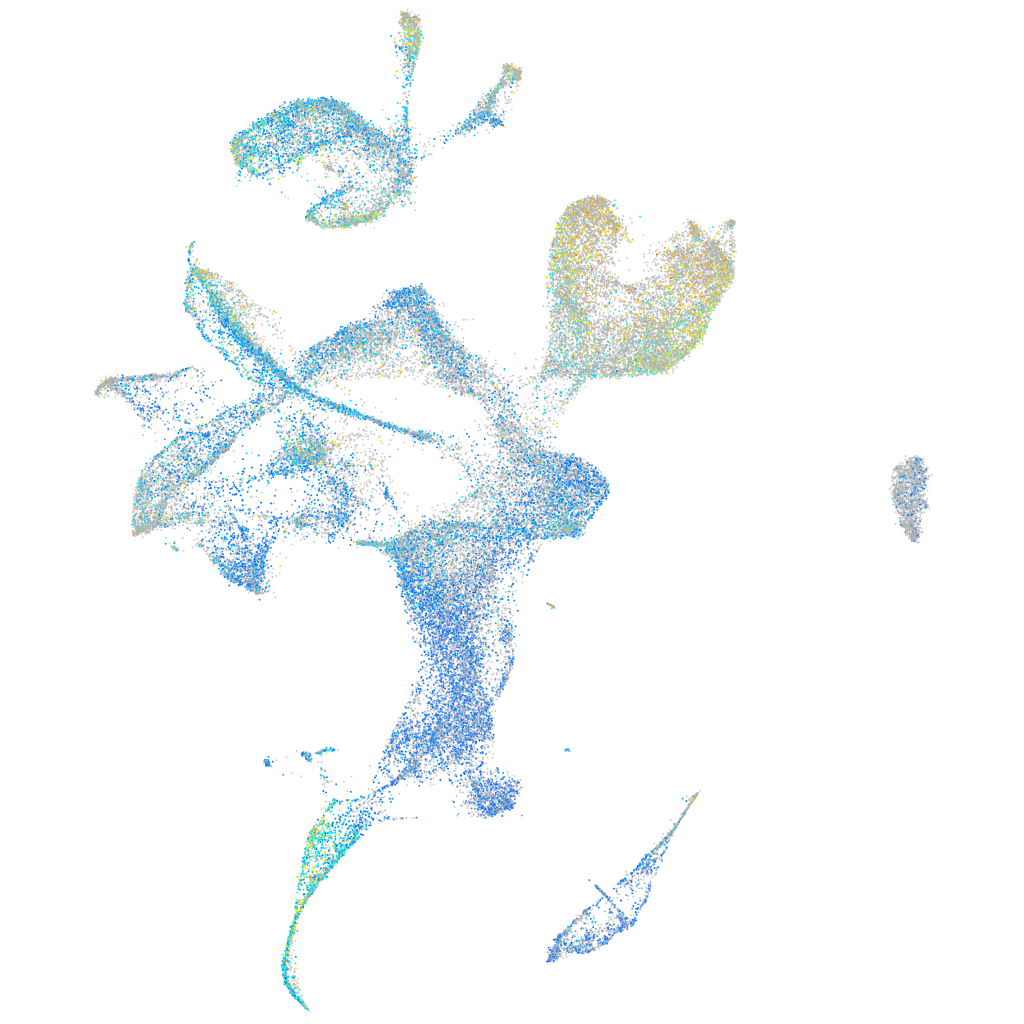"RAB5C, member RAS oncogene family"
ZFIN 
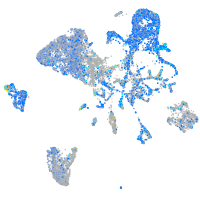
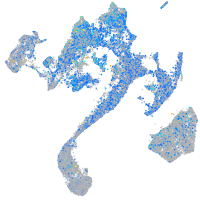
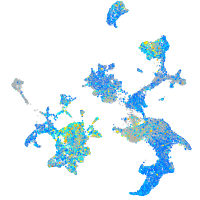
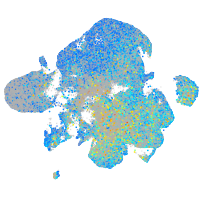
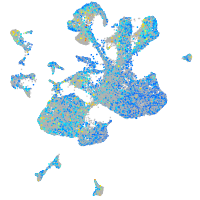
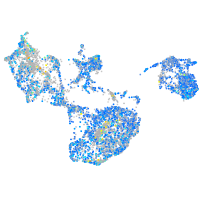
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dct | 0.217 | hmgb2a | -0.084 |
| tyrp1b | 0.214 | si:ch211-222l21.1 | -0.069 |
| gpr143 | 0.213 | stmn1a | -0.068 |
| pmelb | 0.210 | hmga1a | -0.067 |
| pmela | 0.210 | hmgb2b | -0.065 |
| slc45a2 | 0.208 | lig1 | -0.064 |
| tspan10 | 0.201 | shisa2a | -0.064 |
| tyrp1a | 0.200 | fabp7a | -0.062 |
| rab38 | 0.198 | si:ch211-152c2.3 | -0.062 |
| tspan36 | 0.192 | ptmab | -0.061 |
| oca2 | 0.190 | hspb1 | -0.061 |
| fabp11b | 0.188 | pcna | -0.060 |
| cracr2ab | 0.185 | zmp:0000000624 | -0.058 |
| bace2 | 0.185 | hesx1 | -0.056 |
| msnb | 0.183 | ccnd1 | -0.055 |
| rgrb | 0.178 | nasp | -0.055 |
| qdpra | 0.176 | si:dkey-239h2.3 | -0.054 |
| slc24a5 | 0.176 | rrs1 | -0.054 |
| pttg1ipb | 0.174 | tdgf1 | -0.053 |
| vat1 | 0.173 | mcm7 | -0.053 |
| gstp1 | 0.172 | ddx18 | -0.053 |
| agtrap | 0.172 | dkc1 | -0.053 |
| tmem98 | 0.167 | chaf1a | -0.053 |
| tyr | 0.166 | fbl | -0.052 |
| mitfa | 0.166 | mcm6 | -0.052 |
| fgfbp1a | 0.165 | apoc1 | -0.052 |
| tm6sf2 | 0.164 | mki67 | -0.052 |
| mb | 0.162 | sox19a | -0.051 |
| triobpa | 0.162 | si:ch73-1a9.3 | -0.050 |
| rab32a | 0.161 | nop58 | -0.050 |
| zgc:110591 | 0.160 | zgc:110425 | -0.049 |
| stra6 | 0.159 | COX7A2 | -0.049 |
| FP085398.1 | 0.159 | ccne2 | -0.048 |
| rab27a | 0.159 | cyp26a1 | -0.048 |
| sytl2a | 0.158 | gar1 | -0.048 |