"RAB3D, member RAS oncogene family, b"
ZFIN 
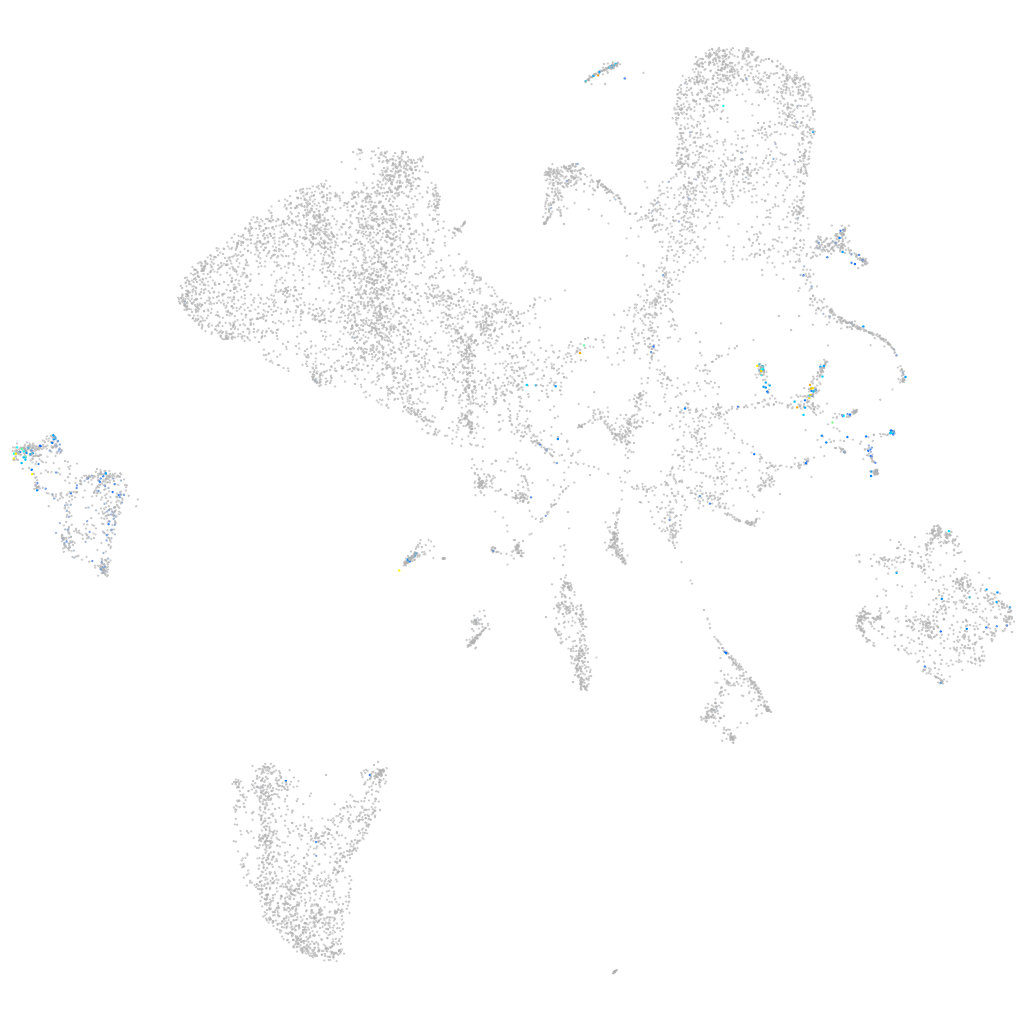
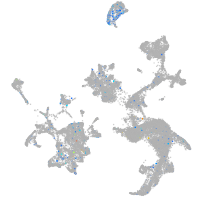
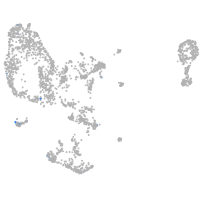
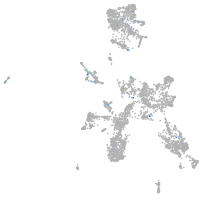
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.260 | gapdh | -0.137 |
| fev | 0.238 | gamt | -0.134 |
| insm1b | 0.216 | ahcy | -0.124 |
| neurod1 | 0.214 | gatm | -0.122 |
| vat1 | 0.209 | bhmt | -0.119 |
| mir7a-1 | 0.209 | fbp1b | -0.115 |
| TCIM (1 of many) | 0.206 | mat1a | -0.114 |
| pax6b | 0.203 | apoa1b | -0.108 |
| mir375-2 | 0.201 | apoa4b.1 | -0.106 |
| c2cd4a | 0.200 | cx32.3 | -0.106 |
| rprmb | 0.198 | apoc2 | -0.105 |
| pax4 | 0.198 | gstt1a | -0.105 |
| gapdhs | 0.195 | afp4 | -0.105 |
| capsla | 0.195 | si:dkey-16p21.8 | -0.104 |
| calm1b | 0.192 | hspe1 | -0.102 |
| pcsk1nl | 0.191 | glud1b | -0.100 |
| zgc:101731 | 0.191 | aqp12 | -0.100 |
| hepacam2 | 0.189 | scp2a | -0.099 |
| cplx2 | 0.186 | gstr | -0.099 |
| si:zfos-2372e4.1 | 0.185 | agxta | -0.099 |
| egr4 | 0.185 | apoa2 | -0.098 |
| vamp2 | 0.185 | agxtb | -0.096 |
| map4l | 0.182 | aldh7a1 | -0.096 |
| CR556712.1 | 0.181 | aldh6a1 | -0.095 |
| gpc1a | 0.180 | tfa | -0.095 |
| cldnh | 0.180 | grhprb | -0.094 |
| kcnk3b | 0.180 | adka | -0.093 |
| trhra | 0.178 | gcshb | -0.093 |
| jph3 | 0.177 | apobb.1 | -0.093 |
| si:ch73-160i9.2 | 0.176 | suclg1 | -0.093 |
| b3gnt7 | 0.174 | nipsnap3a | -0.092 |
| pcdh20 | 0.174 | nupr1b | -0.092 |
| pygma | 0.173 | ftcd | -0.091 |
| cacna1c | 0.172 | pnp4b | -0.091 |
| tmem63c | 0.170 | serpina1 | -0.091 |