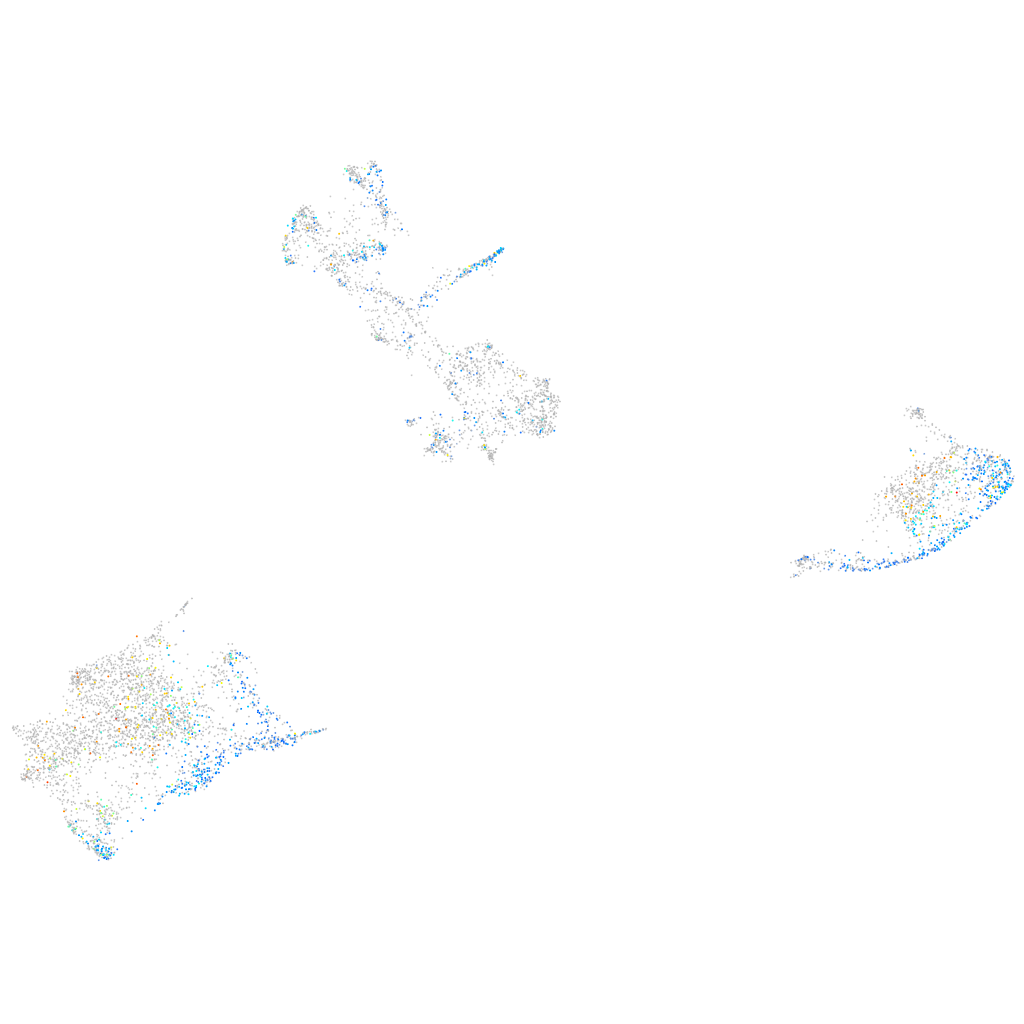
"RAB38c, member of RAS oncogene family"
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 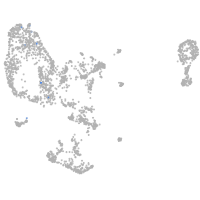 |
neural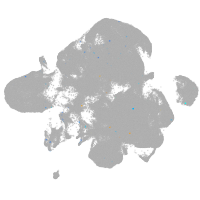 |
spinal cord / glia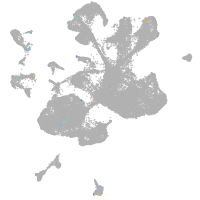 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm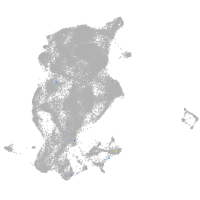 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pah | 0.159 | si:ch211-222l21.1 | -0.108 |
| tspan36 | 0.158 | ptmaa | -0.092 |
| slc3a2a | 0.158 | nova2 | -0.092 |
| tyr | 0.149 | si:ch73-1a9.3 | -0.091 |
| rab34b | 0.148 | mdkb | -0.087 |
| kcnj13 | 0.147 | tuba1c | -0.087 |
| si:zfos-943e10.1 | 0.146 | gpm6aa | -0.086 |
| tspan10 | 0.145 | hmgb1b | -0.085 |
| pttg1ipb | 0.145 | elavl3 | -0.083 |
| mitfa | 0.145 | epb41a | -0.080 |
| slc24a5 | 0.145 | hmgb2b | -0.078 |
| slc37a2 | 0.140 | tmsb | -0.074 |
| rab32a | 0.138 | hmgb3a | -0.074 |
| kita | 0.138 | gng3 | -0.073 |
| SPAG9 | 0.137 | fabp7a | -0.071 |
| qdpra | 0.136 | stmn1b | -0.069 |
| aadac | 0.135 | stmn1a | -0.069 |
| syngr1a | 0.135 | zc4h2 | -0.068 |
| exoc3l1 | 0.134 | si:ch73-281n10.2 | -0.067 |
| pcbd1 | 0.134 | csdc2a | -0.064 |
| tyrp1a | 0.133 | sox11b | -0.064 |
| inka1b | 0.132 | cadm3 | -0.062 |
| opn5 | 0.132 | vamp2 | -0.062 |
| tfap2e | 0.131 | tmeff1b | -0.062 |
| atp11a | 0.131 | myt1b | -0.062 |
| id3 | 0.131 | pcna | -0.062 |
| bace2 | 0.130 | XLOC-003692 | -0.061 |
| lamp1a | 0.130 | CU634008.1 | -0.061 |
| dct | 0.130 | hbbe1.1 | -0.059 |
| mlpha | 0.130 | fez1 | -0.059 |
| gramd2aa | 0.129 | hmgn2 | -0.059 |
| pmela | 0.129 | marcksb | -0.059 |
| pcdh10a | 0.129 | CU467822.1 | -0.059 |
| abracl | 0.128 | h3f3a | -0.059 |
| tdh | 0.128 | tubb5 | -0.059 |