"RAB34, member RAS oncogene family b"
ZFIN 
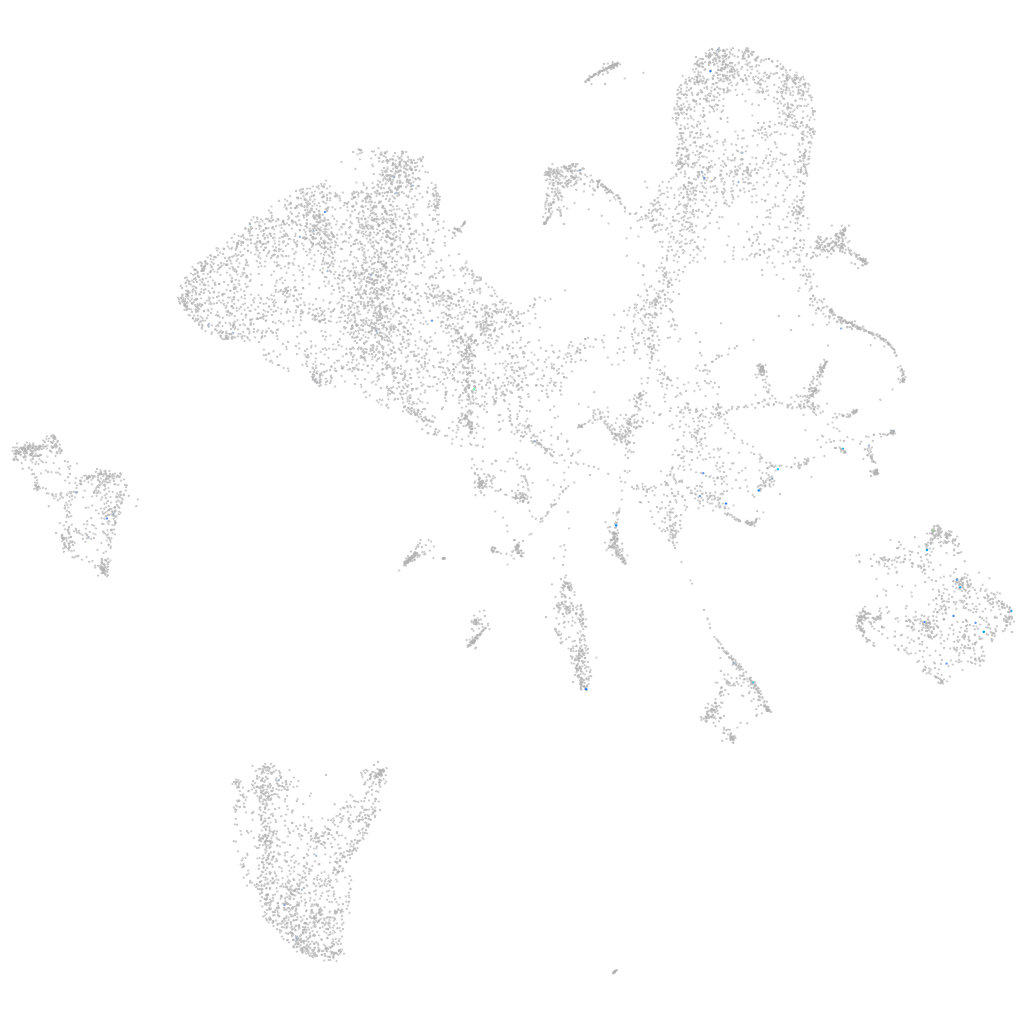
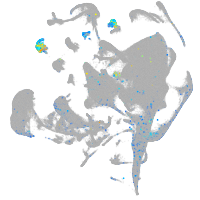
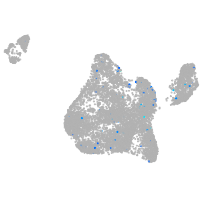
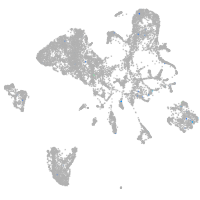
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:zfos-1069f5.1 | 0.329 | atp5fa1 | -0.049 |
| CR545476.1 | 0.289 | rpl37 | -0.042 |
| BX469910.2 | 0.252 | zgc:114188 | -0.040 |
| SH3RF3 | 0.231 | ahcy | -0.039 |
| CT737129.1 | 0.218 | rps10 | -0.038 |
| slc2a15a | 0.214 | atp5f1c | -0.037 |
| tmx3b | 0.214 | ndufb7 | -0.037 |
| FOXN2 | 0.212 | aldob | -0.037 |
| itln1 | 0.207 | zgc:158463 | -0.036 |
| pea15 | 0.204 | mpc2 | -0.036 |
| si:dkeyp-51f12.3 | 0.200 | atp5pf | -0.036 |
| LOC110438679 | 0.186 | mt-atp6 | -0.036 |
| spi1b | 0.170 | rps17 | -0.035 |
| bscl2l | 0.169 | eef1da | -0.034 |
| LOC103910470 | 0.167 | atp5f1d | -0.034 |
| CR932000.1 | 0.163 | sod2 | -0.033 |
| ednrba | 0.160 | COX3 | -0.033 |
| tmem130 | 0.155 | sod1 | -0.033 |
| CU929150.2 | 0.150 | BX908782.3 | -0.033 |
| XLOC-042977 | 0.147 | zgc:92744 | -0.033 |
| tal1 | 0.146 | gapdh | -0.032 |
| XLOC-019062 | 0.143 | eno3 | -0.032 |
| LOC101885586 | 0.142 | tpi1b | -0.032 |
| flnca | 0.140 | gstt1a | -0.032 |
| ackr3a | 0.139 | aldh7a1 | -0.032 |
| BX088688.3 | 0.136 | nme2b.1 | -0.032 |
| dio3a | 0.136 | gamt | -0.032 |
| myot | 0.135 | aldh6a1 | -0.032 |
| BX004774.2 | 0.134 | prdx2 | -0.031 |
| si:dkey-21o22.2 | 0.132 | atp5mc1 | -0.031 |
| pax7b | 0.130 | ndufb10 | -0.031 |
| LOC108183291 | 0.128 | acadl | -0.031 |
| LOC108183690 | 0.126 | suclg1 | -0.031 |
| LOC101883560 | 0.126 | cox6b1 | -0.031 |
| plppr2b | 0.126 | ndufa2 | -0.031 |