R3H domain and coiled-coil containing 1-like
ZFIN 
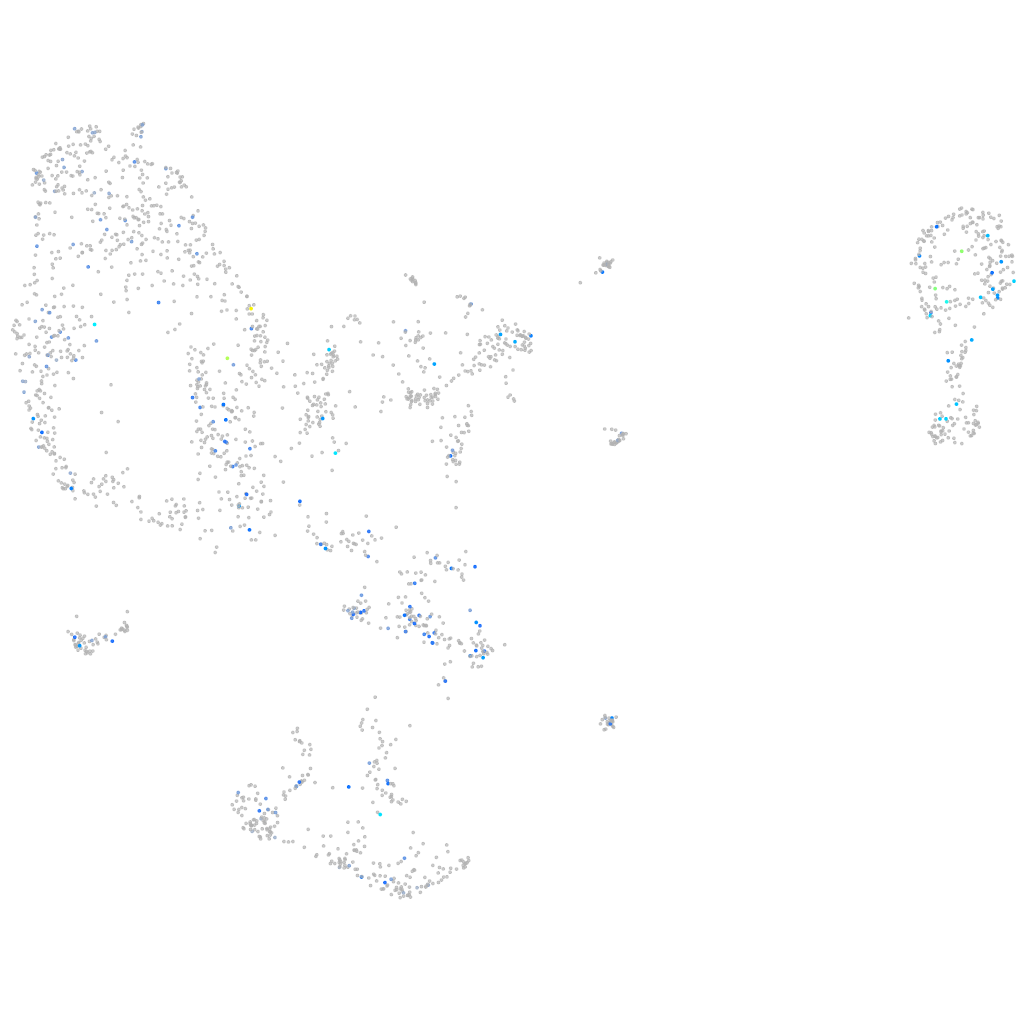
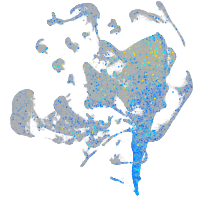
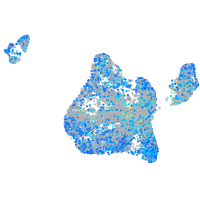
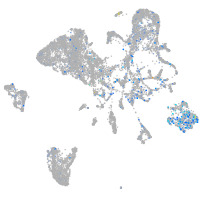
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01048053.1 | 0.199 | si:ch211-139a5.9 | -0.084 |
| RF00213 | 0.199 | pnp6 | -0.082 |
| CT025913.1 | 0.197 | ndufa7 | -0.076 |
| XLOC-042669 | 0.179 | gapdh | -0.075 |
| alox5b.3 | 0.175 | ndufa9a | -0.075 |
| gig2h | 0.167 | COX3 | -0.075 |
| slc4a5a | 0.160 | gstk1 | -0.074 |
| asz1 | 0.160 | aldob | -0.074 |
| RF00430 | 0.154 | pvalb1 | -0.072 |
| BX908753.1 | 0.152 | mt-atp8 | -0.071 |
| BX890559.1 | 0.150 | elovl5 | -0.069 |
| CR854838.1 | 0.150 | tmem256 | -0.068 |
| XLOC-003625 | 0.145 | cubn | -0.068 |
| CT025912.1 | 0.144 | glud1b | -0.068 |
| CU683879.2 | 0.144 | akr7a3 | -0.067 |
| LOC110438610 | 0.143 | cdaa | -0.067 |
| u2af1 | 0.142 | aco1 | -0.066 |
| LOC101884617 | 0.141 | atp6v1e1b | -0.066 |
| LOC101882460 | 0.141 | mt-co1 | -0.065 |
| fsip1.1 | 0.140 | cox5ab | -0.065 |
| lpar6a | 0.140 | eno3 | -0.065 |
| XLOC-008823 | 0.139 | mt-nd1 | -0.065 |
| si:dkey-157e10.8 | 0.137 | ndufa4l | -0.065 |
| lrrc28 | 0.136 | tpma | -0.065 |
| fgf2 | 0.135 | mt-nd2 | -0.064 |
| LOC103911969 | 0.135 | galnt14 | -0.064 |
| ackr4a | 0.134 | cox6b1 | -0.064 |
| irgf1 | 0.132 | pgam1a | -0.064 |
| htatsf1 | 0.126 | DHDH | -0.064 |
| XLOC-039363 | 0.125 | sod2 | -0.064 |
| BX957292.1 | 0.124 | cebpd | -0.064 |
| hipk1a | 0.124 | icn | -0.064 |
| CR394552.1 | 0.124 | hadh | -0.063 |
| tmem62 | 0.123 | vat1 | -0.063 |
| AL929210.2 | 0.123 | aldh6a1 | -0.063 |