"QKI, KH domain containing, RNA binding 2"
ZFIN 
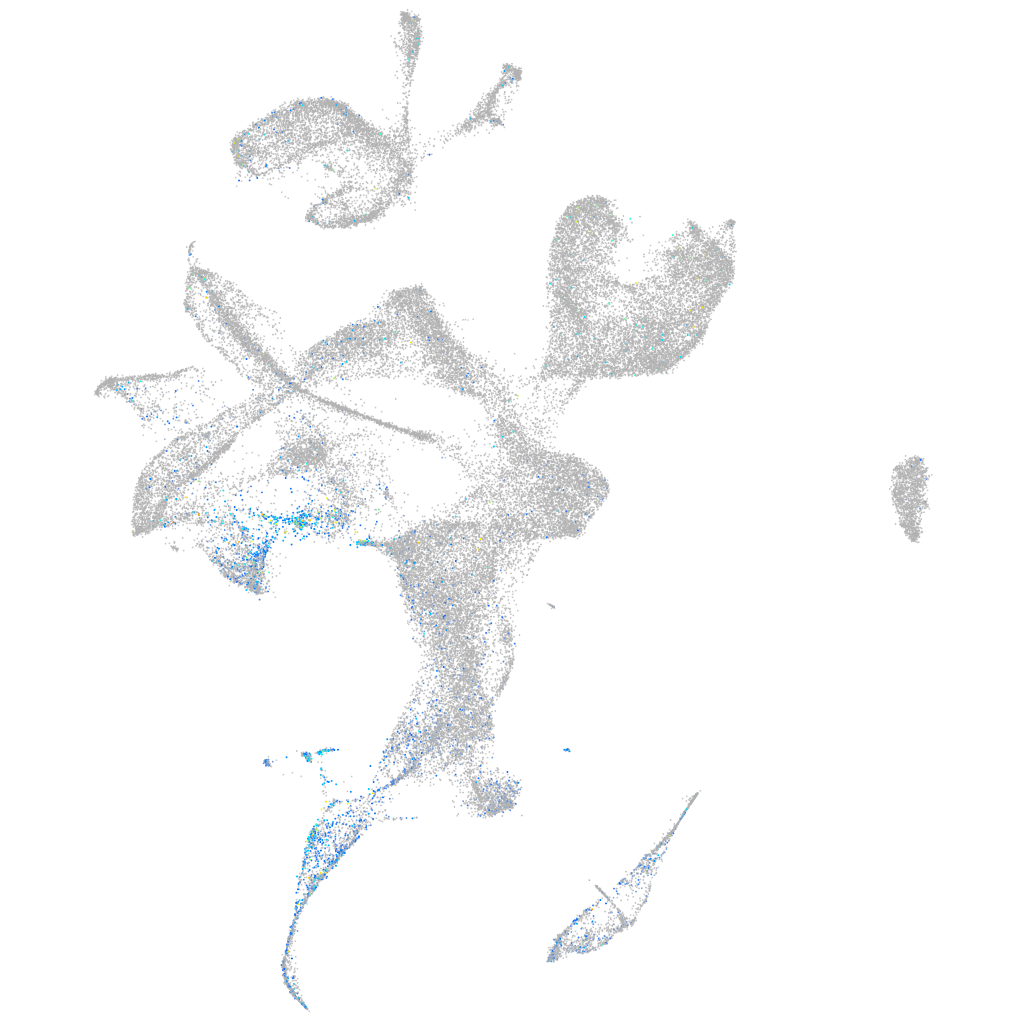
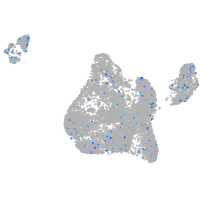
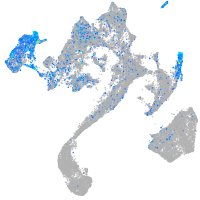
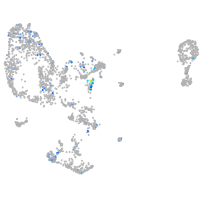
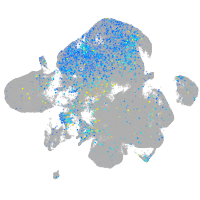
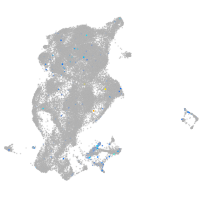
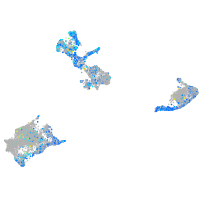
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sparc | 0.237 | ptmab | -0.165 |
| cldn19 | 0.230 | si:ch73-1a9.3 | -0.115 |
| cd63 | 0.228 | hmgn6 | -0.100 |
| hepacama | 0.219 | hmgb1a | -0.080 |
| slc3a2a | 0.219 | ptmaa | -0.079 |
| cx43 | 0.215 | h2afva | -0.078 |
| atp1a1b | 0.211 | cadm3 | -0.075 |
| si:ch211-66e2.5 | 0.205 | anp32e | -0.071 |
| col4a5 | 0.205 | hnrnpaba | -0.071 |
| col4a6 | 0.203 | hmgn2 | -0.071 |
| tspan36 | 0.200 | epb41a | -0.071 |
| slc45a2 | 0.200 | si:ch211-137a8.4 | -0.070 |
| wls | 0.200 | neurod4 | -0.070 |
| fstl1b | 0.200 | gpm6aa | -0.068 |
| pmela | 0.196 | foxg1b | -0.066 |
| gstp1 | 0.194 | cirbpb | -0.066 |
| atp1b4 | 0.193 | tuba1c | -0.063 |
| dct | 0.192 | rorb | -0.063 |
| tyrp1b | 0.191 | smarce1 | -0.063 |
| rnaseka | 0.189 | sumo3a | -0.061 |
| ppap2d | 0.187 | hmga1a | -0.061 |
| rab38 | 0.186 | hnrnpa0a | -0.060 |
| igfbp7 | 0.182 | h3f3d | -0.060 |
| ldlrap1a | 0.182 | scrt2 | -0.059 |
| cyp2ad3 | 0.181 | tp53inp2 | -0.058 |
| bace2 | 0.180 | hmgb1b | -0.058 |
| slc1a3b | 0.179 | marcksl1b | -0.057 |
| tspan10 | 0.179 | marcksb | -0.056 |
| sytl2b | 0.178 | sinhcafl | -0.056 |
| tyrp1a | 0.178 | si:ch211-288g17.3 | -0.056 |
| qdpra | 0.178 | tuba1a | -0.055 |
| hmga2 | 0.177 | btg1 | -0.055 |
| pmelb | 0.177 | vsx1 | -0.055 |
| pcbd1 | 0.176 | ndrg4 | -0.055 |
| oca2 | 0.176 | insm1a | -0.055 |