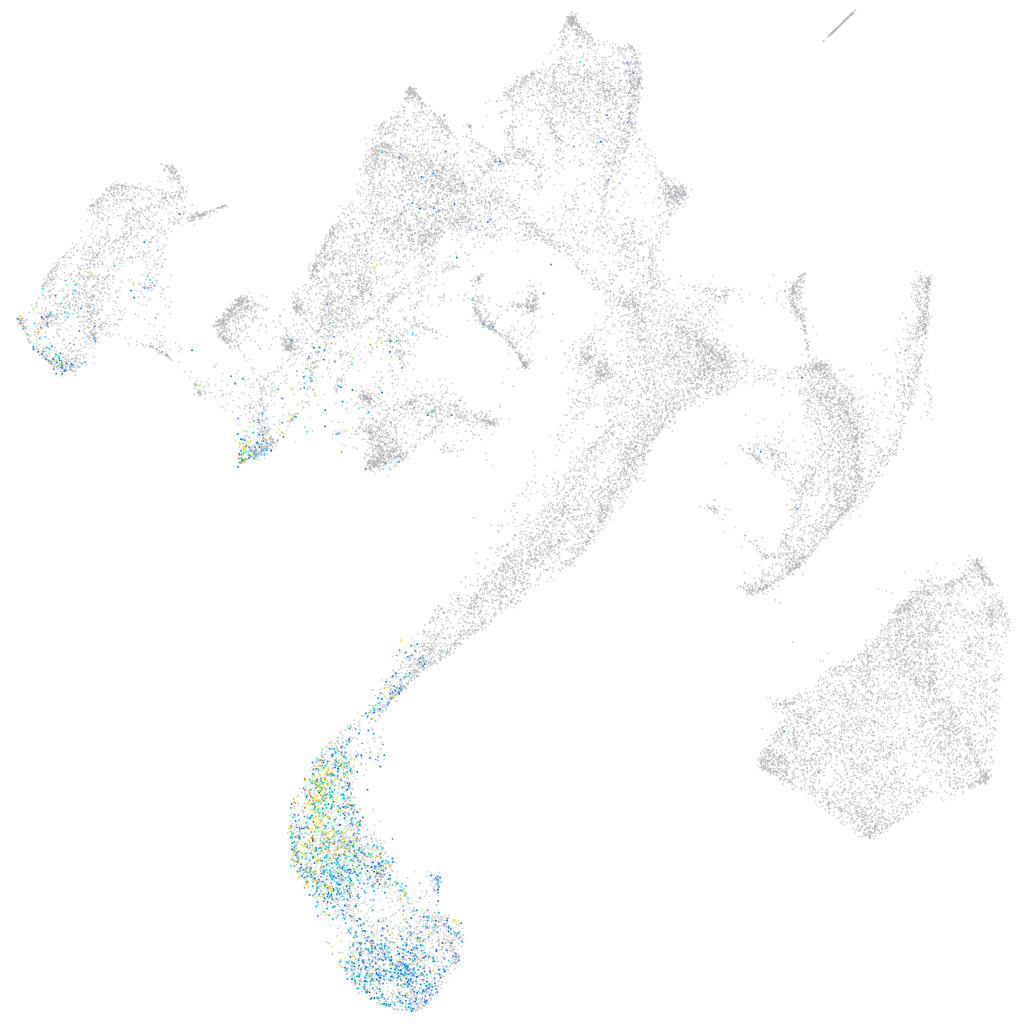
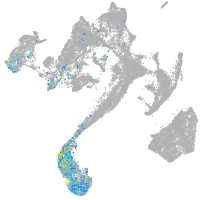
parvalbumin 3
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tnni2a.4 | 0.477 | hsp90ab1 | -0.380 |
| myoz1b | 0.470 | h3f3d | -0.376 |
| tnnt3a | 0.468 | cirbpb | -0.349 |
| tnnt3b | 0.467 | pabpc1a | -0.346 |
| mylpfb | 0.467 | h2afvb | -0.336 |
| casq1a | 0.463 | khdrbs1a | -0.334 |
| pvalb1 | 0.460 | ran | -0.328 |
| myom2a | 0.460 | fthl27 | -0.322 |
| si:ch73-367p23.2 | 0.458 | hnrnpaba | -0.322 |
| pvalb2 | 0.457 | hmga1a | -0.316 |
| mylz3 | 0.455 | si:ch73-1a9.3 | -0.314 |
| myhz1.3 | 0.448 | actb2 | -0.313 |
| CABZ01061524.1 | 0.440 | ubc | -0.312 |
| hhatla | 0.436 | naca | -0.311 |
| mybpc2b | 0.436 | hmgb2b | -0.305 |
| slc25a4 | 0.436 | cirbpa | -0.303 |
| ryr3 | 0.435 | hmgn2 | -0.296 |
| mylpfa | 0.432 | hnrnpabb | -0.295 |
| myl1 | 0.430 | cfl1 | -0.295 |
| acta1b | 0.422 | hnrnpa0b | -0.292 |
| actn3a | 0.422 | hmgb2a | -0.291 |
| tnnc2 | 0.420 | syncrip | -0.288 |
| tpma | 0.419 | ptmab | -0.288 |
| XLOC-001975 | 0.416 | si:ch211-222l21.1 | -0.285 |
| smyd1a | 0.415 | setb | -0.285 |
| ckma | 0.414 | hmgn7 | -0.283 |
| myom1a | 0.414 | snrpf | -0.282 |
| cdnf | 0.410 | si:ch73-281n10.2 | -0.281 |
| neb | 0.410 | hmgn6 | -0.281 |
| ckmb | 0.409 | cbx3a | -0.277 |
| XLOC-006515 | 0.409 | eef1a1l1 | -0.275 |
| tmod4 | 0.407 | rbm8a | -0.270 |
| XLOC-025819 | 0.407 | snrpb | -0.264 |
| myhz2 | 0.405 | tuba8l4 | -0.263 |
| si:ch211-255p10.3 | 0.404 | snrpd1 | -0.262 |