purine-rich element binding protein Aa
ZFIN 
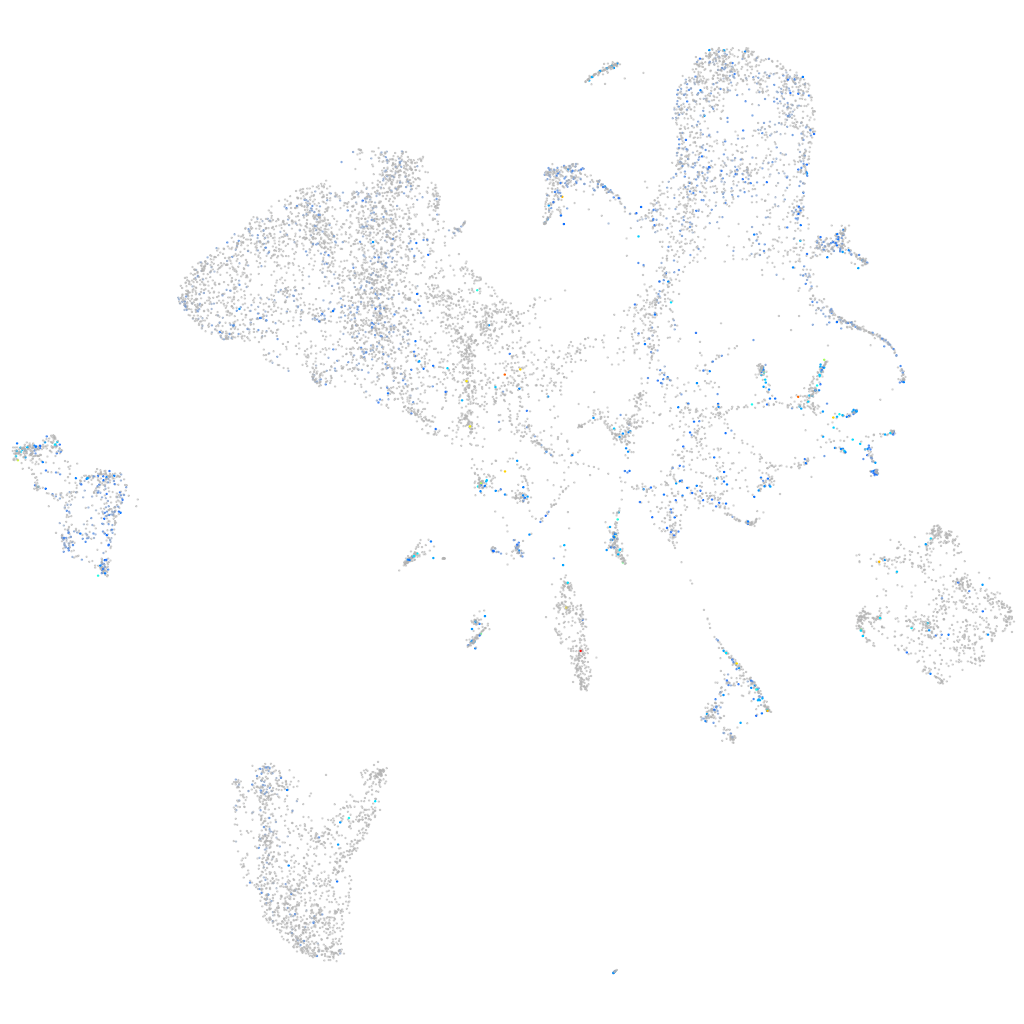
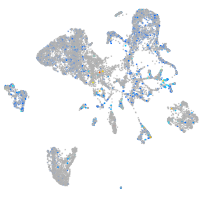
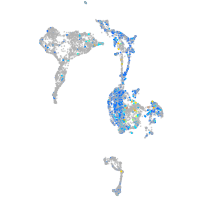
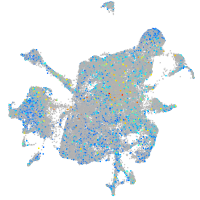
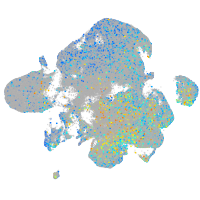
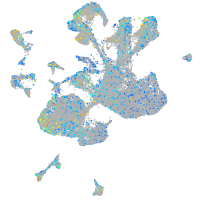
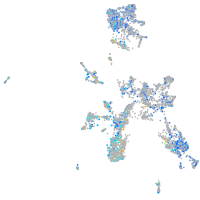
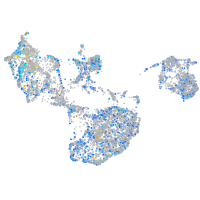
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.176 | bhmt | -0.135 |
| calm1b | 0.171 | gamt | -0.128 |
| gnb1a | 0.170 | gatm | -0.123 |
| calm1a | 0.163 | aqp12 | -0.121 |
| capns1b | 0.162 | apoc1 | -0.113 |
| ier2b | 0.162 | gapdh | -0.113 |
| vat1 | 0.161 | mat1a | -0.113 |
| scg3 | 0.156 | agxtb | -0.113 |
| ppiab | 0.156 | apoa1b | -0.101 |
| CR383676.1 | 0.156 | apoa2 | -0.100 |
| elovl1b | 0.155 | rbp2b | -0.099 |
| arpc3 | 0.155 | fetub | -0.099 |
| ccni | 0.153 | pnp4b | -0.099 |
| rasd4 | 0.151 | ahcy | -0.098 |
| cd63 | 0.151 | ttc36 | -0.096 |
| scgn | 0.150 | apom | -0.095 |
| kdm6bb | 0.148 | serpina1 | -0.094 |
| rprmb | 0.147 | serpina1l | -0.094 |
| fhl3b | 0.147 | grhprb | -0.094 |
| arhgdia | 0.146 | kng1 | -0.094 |
| tmem59 | 0.145 | hao1 | -0.093 |
| calm3b | 0.144 | ces2 | -0.093 |
| ptmaa | 0.142 | fabp10a | -0.093 |
| btg1 | 0.142 | zgc:123103 | -0.092 |
| lasp1 | 0.141 | LOC110437731 | -0.090 |
| cap1 | 0.140 | rbp4 | -0.090 |
| si:ch1073-429i10.3.1 | 0.139 | ambp | -0.090 |
| GCA | 0.139 | fgb | -0.089 |
| si:dkey-112e7.2 | 0.139 | gc | -0.089 |
| pax6b | 0.139 | uox | -0.089 |
| dkk3b | 0.138 | fgg | -0.088 |
| zgc:153867 | 0.137 | tfa | -0.088 |
| rap1b | 0.137 | si:dkey-16p21.8 | -0.087 |
| ubc | 0.136 | ttr | -0.086 |
| rassf4a | 0.136 | ftcd | -0.086 |