peptidyl-tRNA hydrolase 2
ZFIN 
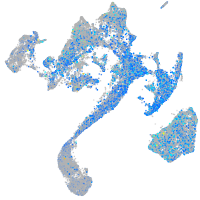
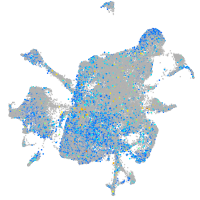
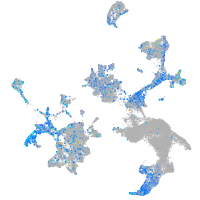
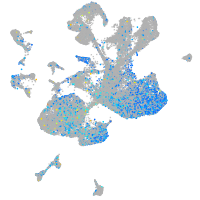
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tomm40l | 0.209 | rplp2 | -0.091 |
| dut | 0.195 | dap1b | -0.089 |
| alyref | 0.188 | tinagl1 | -0.079 |
| lsm7 | 0.187 | CR383676.1 | -0.079 |
| mcm7 | 0.182 | rtn1a | -0.077 |
| vcana | 0.182 | icn2 | -0.075 |
| eif4a1a | 0.181 | hmha1a | -0.074 |
| snrpd1 | 0.181 | mt-co2 | -0.073 |
| ptges3b | 0.180 | map1aa | -0.073 |
| psma6a | 0.180 | rps29 | -0.072 |
| h2afvb | 0.180 | tspo | -0.072 |
| erh | 0.180 | krt8 | -0.072 |
| psmb1 | 0.178 | si:ch211-139a5.9 | -0.072 |
| sf3a2 | 0.176 | cldn8 | -0.071 |
| psmd7 | 0.175 | pvalb1 | -0.071 |
| ewsr1b | 0.175 | atp1b3b | -0.070 |
| snrpb | 0.174 | nsg2 | -0.070 |
| snrpg | 0.173 | pleca | -0.069 |
| eif4a3 | 0.173 | rpl3 | -0.069 |
| snrpd2 | 0.172 | aldob | -0.068 |
| psma3 | 0.170 | tmsb | -0.067 |
| BX469925.3 | 0.169 | basp1 | -0.067 |
| crtap | 0.169 | sat1a.1 | -0.067 |
| psma5 | 0.169 | elavl3 | -0.067 |
| tma7 | 0.169 | mt-atp8 | -0.066 |
| psmb7 | 0.168 | elavl4 | -0.065 |
| slirp | 0.168 | vdrb | -0.065 |
| cdkn2aipnl | 0.168 | mt-nd3 | -0.065 |
| sap18 | 0.168 | mt-co1 | -0.065 |
| h2afva | 0.167 | cnrip1a | -0.064 |
| tuba8l4 | 0.167 | syn2a | -0.064 |
| snrpf | 0.167 | tspan9a | -0.064 |
| khdrbs1a | 0.166 | gchfr | -0.064 |
| hnrnpabb | 0.166 | capga | -0.063 |
| sumo3a | 0.166 | ptgir | -0.063 |