protein tyrosine phosphatase receptor type Q
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line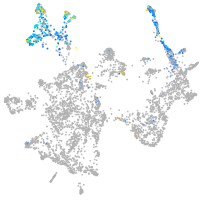 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nphs2 | 0.632 | pmp22a | -0.032 |
| si:ch73-205h11.1 | 0.524 | col5a1 | -0.030 |
| ptprh | 0.494 | col1a2 | -0.025 |
| nphs1 | 0.466 | krt18a.1 | -0.025 |
| itgb6 | 0.338 | fkbp7 | -0.023 |
| naalad2 | 0.336 | si:dkey-261h17.1 | -0.022 |
| fam49ba | 0.323 | krt18b | -0.022 |
| iqgap2 | 0.295 | LO018188.1 | -0.022 |
| il2rb | 0.251 | col2a1b | -0.021 |
| sh3d19 | 0.247 | krt8 | -0.021 |
| col4a4 | 0.237 | marcksl1a | -0.021 |
| sema7a | 0.219 | fmoda | -0.021 |
| lrmp | 0.215 | fibina | -0.020 |
| itga3b | 0.211 | tuba8l3 | -0.020 |
| map3k15 | 0.203 | hmga2 | -0.020 |
| wt1b | 0.201 | twist1b | -0.020 |
| col4a3 | 0.197 | pdgfra | -0.020 |
| wnt2ba | 0.189 | col1a1a | -0.020 |
| kirrel1b | 0.184 | ahcy | -0.020 |
| pvalb7 | 0.182 | bhmt | -0.019 |
| podxl | 0.181 | lpar1 | -0.019 |
| b3gat2 | 0.180 | twist1a | -0.019 |
| vegfba | 0.177 | foxp4 | -0.019 |
| ankdd1b | 0.172 | rarga | -0.019 |
| cnksr1 | 0.172 | tmem119b | -0.019 |
| magi2a | 0.166 | CU929237.1 | -0.019 |
| tagln3b | 0.164 | dcn | -0.019 |
| efhd1 | 0.163 | col6a1 | -0.018 |
| LOC110439775 | 0.162 | prrx1a | -0.018 |
| ppp4r4 | 0.156 | cd248b | -0.018 |
| pip5k1ca | 0.154 | pleca | -0.018 |
| lama5 | 0.151 | col12a1a | -0.018 |
| ripply1 | 0.146 | fstl1b | -0.018 |
| vegfab | 0.144 | six1a | -0.017 |
| XLOC-032129 | 0.143 | mab21l2 | -0.017 |




