protein tyrosine phosphatase non-receptor type 18
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 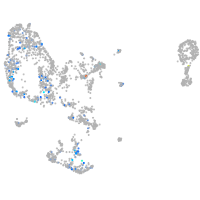 |
neural |
spinal cord / glia |
eye |
taste / olfactory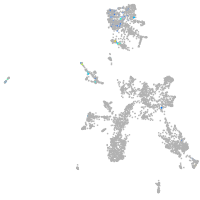 |
otic / lateral line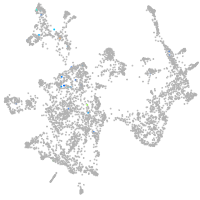 |
epidermis |
periderm |
fin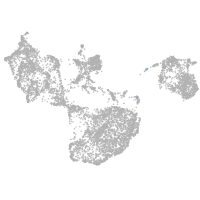 |
ionocytes / mucous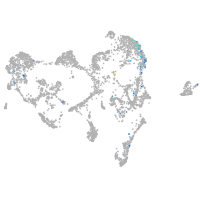 |
pigment cells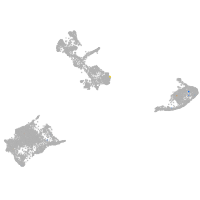 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| vill | 0.110 | nova2 | -0.033 |
| cdh17 | 0.107 | gpm6aa | -0.032 |
| prr15la | 0.107 | hmgb1b | -0.031 |
| cldnh | 0.101 | tuba1c | -0.030 |
| si:ch73-185c24.2 | 0.101 | mdkb | -0.029 |
| gucy2c | 0.100 | tuba1a | -0.028 |
| lrrc66 | 0.100 | elavl3 | -0.027 |
| eps8l3b | 0.096 | gpm6ab | -0.027 |
| eps8l3a | 0.092 | tubb5 | -0.026 |
| tmprss2 | 0.092 | marcksl1a | -0.026 |
| atp1a1a.4 | 0.091 | rtn1a | -0.025 |
| grn1 | 0.091 | stmn1b | -0.025 |
| si:dkey-194e6.1 | 0.091 | hmgb3a | -0.024 |
| pdzk1 | 0.090 | tmeff1b | -0.024 |
| clic5b | 0.090 | cadm3 | -0.023 |
| cdaa | 0.089 | cspg5a | -0.023 |
| ggt1b | 0.089 | fam168a | -0.023 |
| tmc4 | 0.089 | hnrnpa0a | -0.023 |
| amn | 0.088 | marcksb | -0.023 |
| cldnc | 0.088 | si:ch211-137a8.4 | -0.023 |
| bin2a | 0.087 | atp6v0cb | -0.022 |
| ezra | 0.087 | si:dkey-276j7.1 | -0.022 |
| hnf4g | 0.087 | celf2 | -0.021 |
| si:ch1073-443f11.2 | 0.087 | fabp3 | -0.021 |
| si:dkey-121j17.5 | 0.087 | mdka | -0.021 |
| si:ch211-139a5.9 | 0.087 | zc4h2 | -0.021 |
| cox6a2 | 0.086 | cd99l2 | -0.020 |
| hnf1ba | 0.086 | gng3 | -0.020 |
| st3gal7 | 0.086 | nat8l | -0.020 |
| cldn8 | 0.085 | nkain1 | -0.020 |
| pou2f3 | 0.084 | rtn1b | -0.020 |
| tspan35 | 0.084 | si:ch211-133n4.4 | -0.020 |
| CU682777.2 | 0.083 | si:ch211-288g17.3 | -0.020 |
| lad1 | 0.082 | sncb | -0.020 |
| malb | 0.082 | dbn1 | -0.019 |




