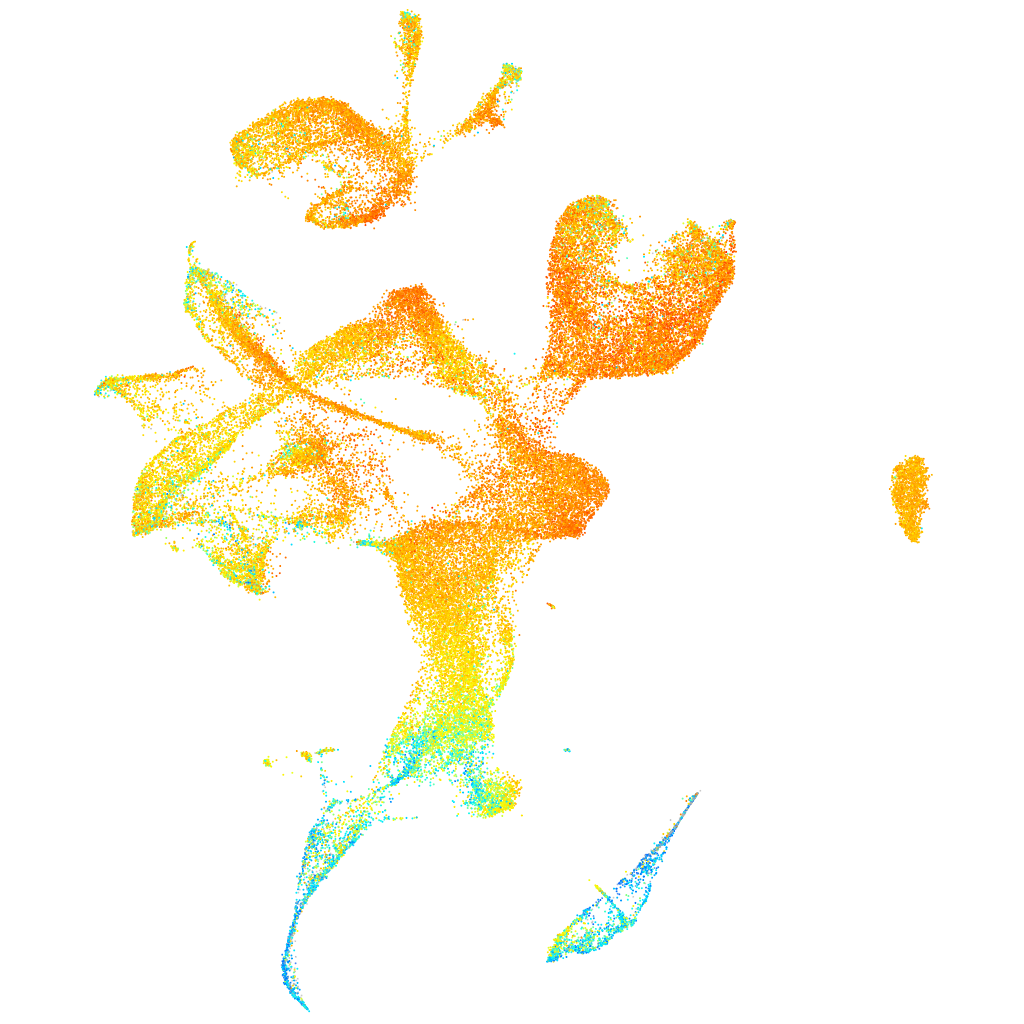prothymosin alpha b
ZFIN 
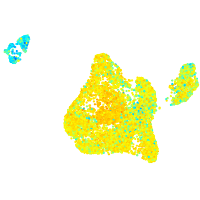
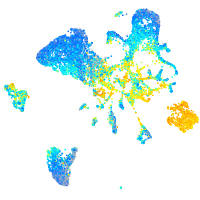
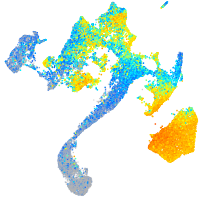
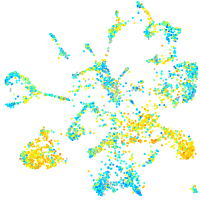
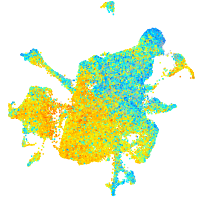
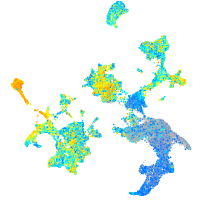
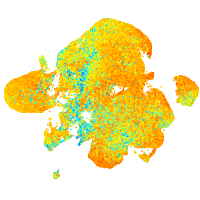
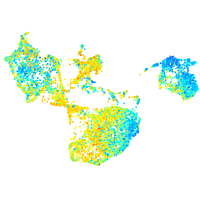
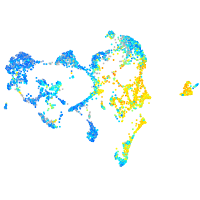
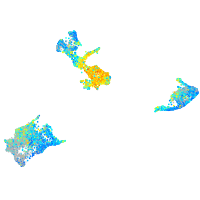
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgn6 | 0.497 | fabp11a | -0.466 |
| si:ch73-1a9.3 | 0.477 | tspan36 | -0.424 |
| hmgb1a | 0.440 | pcbd1 | -0.372 |
| ptmaa | 0.423 | col4a5 | -0.368 |
| h2afva | 0.380 | pmela | -0.365 |
| hmgn2 | 0.379 | dct | -0.365 |
| h3f3d | 0.367 | col4a6 | -0.364 |
| hnrnpaba | 0.358 | cx43 | -0.363 |
| marcksl1b | 0.354 | rgrb | -0.357 |
| cirbpb | 0.351 | tyrp1b | -0.355 |
| ubc | 0.341 | pmelb | -0.352 |
| hmgb1b | 0.334 | sparc | -0.348 |
| gpm6aa | 0.330 | crybb1 | -0.345 |
| neurod4 | 0.314 | atp1b1a | -0.345 |
| tp53inp2 | 0.309 | eef1da | -0.341 |
| btg1 | 0.308 | cryba4 | -0.339 |
| chd4a | 0.308 | cryba2a | -0.338 |
| tuba1a | 0.302 | cryba1l1 | -0.335 |
| hnrnpa0a | 0.297 | slc45a2 | -0.332 |
| h3f3a | 0.295 | pttg1ipb | -0.332 |
| cadm3 | 0.292 | tyrp1a | -0.328 |
| nova2 | 0.291 | cryba1b | -0.327 |
| epb41a | 0.289 | oca2 | -0.327 |
| smarce1 | 0.288 | slc3a2a | -0.326 |
| gpm6ab | 0.286 | cryba2b | -0.325 |
| bcl2l10 | 0.285 | igfbp7 | -0.325 |
| scrt2 | 0.284 | gstp1 | -0.325 |
| h3f3c | 0.282 | gpr143 | -0.323 |
| marcksl1a | 0.279 | bace2 | -0.321 |
| vsx1 | 0.278 | lim2.4 | -0.321 |
| tuba1c | 0.272 | stra6 | -0.321 |
| h2afvb | 0.267 | lpl | -0.319 |
| hmgb3a | 0.267 | mipb | -0.318 |
| si:ch211-288g17.3 | 0.262 | crygn2 | -0.317 |
| anp32e | 0.257 | crybb1l1 | -0.317 |