protein tyrosine kinase 7a
ZFIN 
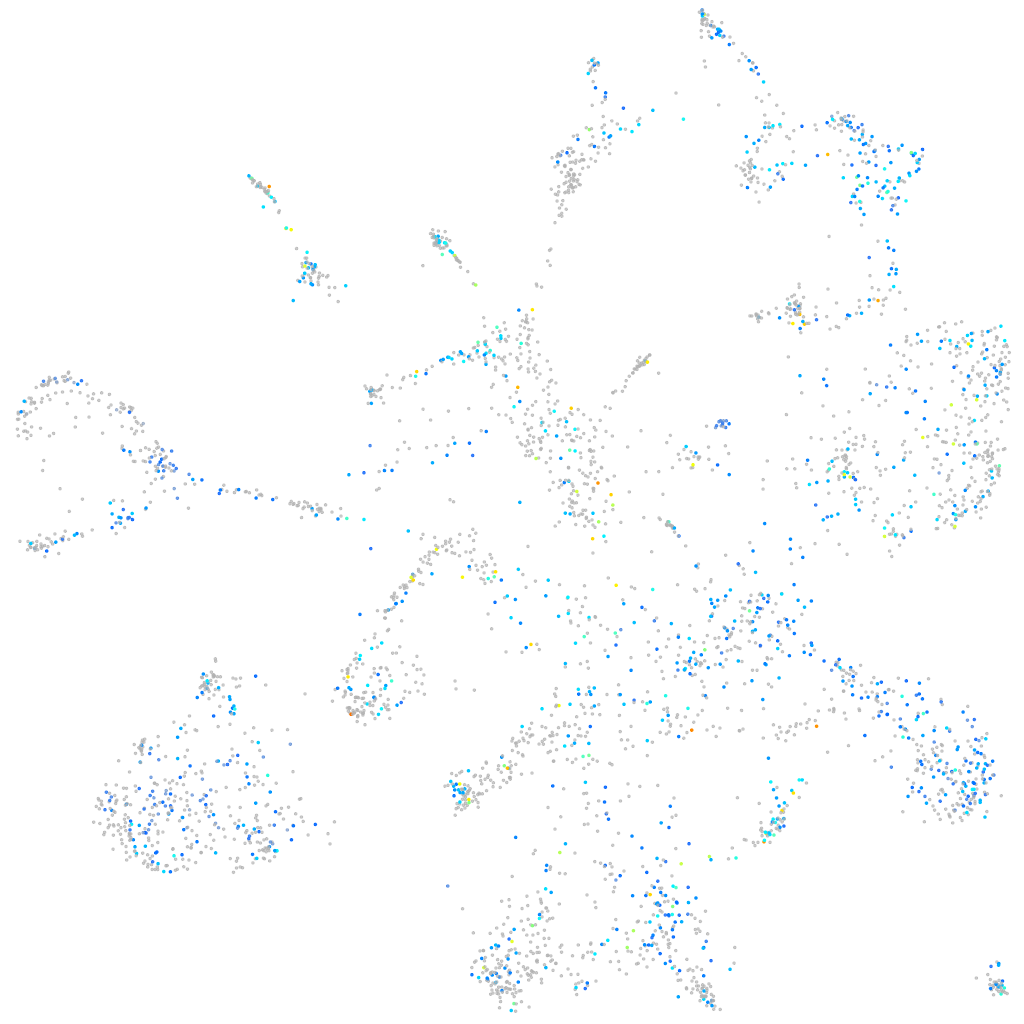
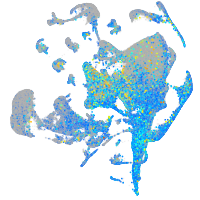
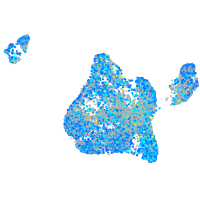
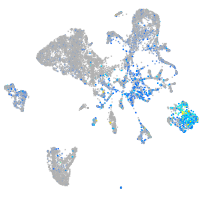
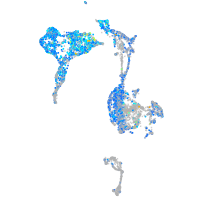
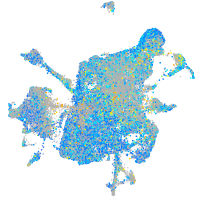
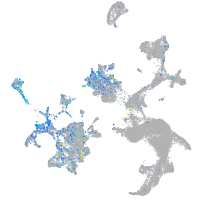
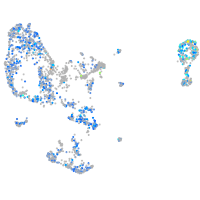
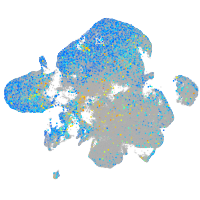
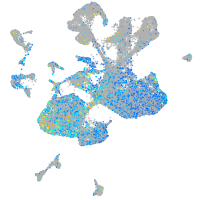
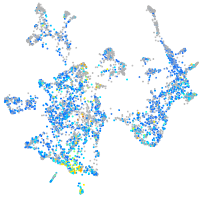
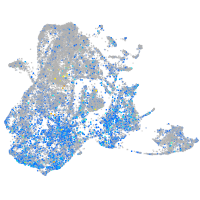
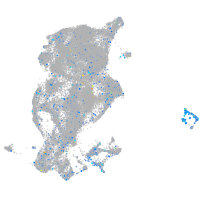
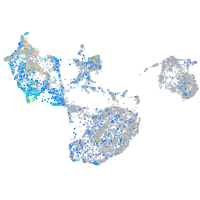
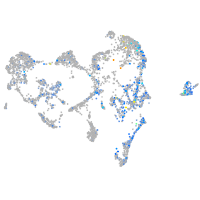
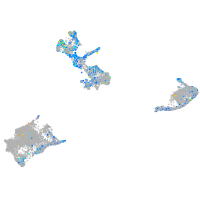
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| bambia | 0.162 | rbp4 | -0.095 |
| calr3b | 0.135 | ifitm1 | -0.084 |
| hnrnpa0l | 0.129 | ctrb1 | -0.078 |
| flot2a | 0.128 | ak1 | -0.075 |
| flot1b | 0.122 | gapdhs | -0.074 |
| h2afva | 0.121 | apoa2 | -0.074 |
| pfdn4 | 0.120 | pfkpa | -0.073 |
| tmem88b | 0.117 | tagln | -0.070 |
| ptbp1a | 0.116 | fbp2 | -0.070 |
| selenof | 0.116 | CU914776.1 | -0.069 |
| tspan7 | 0.116 | prss1 | -0.068 |
| khdrbs1a | 0.114 | pkma | -0.067 |
| hnrnpaba | 0.114 | krt91 | -0.066 |
| lmnb1 | 0.113 | gngt2b | -0.065 |
| msna | 0.113 | CELA1 (1 of many) | -0.064 |
| dad1 | 0.112 | pgk1 | -0.064 |
| si:dkey-261h17.1 | 0.112 | aldocb | -0.063 |
| lin28a | 0.110 | acta2 | -0.063 |
| tnni1b | 0.110 | aldob | -0.062 |
| mdka | 0.110 | pgam1a | -0.060 |
| calr | 0.110 | bgna | -0.059 |
| pdia3 | 0.110 | apoa1b | -0.059 |
| pgrmc1 | 0.110 | loxa | -0.058 |
| fzd7a | 0.109 | synm | -0.058 |
| hnrnpa0b | 0.109 | eno1a | -0.058 |
| jam2b | 0.109 | prss59.1 | -0.058 |
| bmp5 | 0.108 | pvalb2 | -0.058 |
| sec11a | 0.107 | slc16a3 | -0.057 |
| hmga2 | 0.107 | loxl5b | -0.057 |
| fxyd6l | 0.107 | htra1a | -0.057 |
| alpi.1 | 0.107 | prss59.2 | -0.056 |
| crb2a | 0.106 | cnn1b | -0.056 |
| atf4b | 0.106 | XLOC-025423 | -0.056 |
| hmgb2a | 0.105 | cox8b | -0.056 |
| hdac1 | 0.105 | si:dkey-57k2.6 | -0.055 |