polypyrimidine tract binding protein 2a
ZFIN 
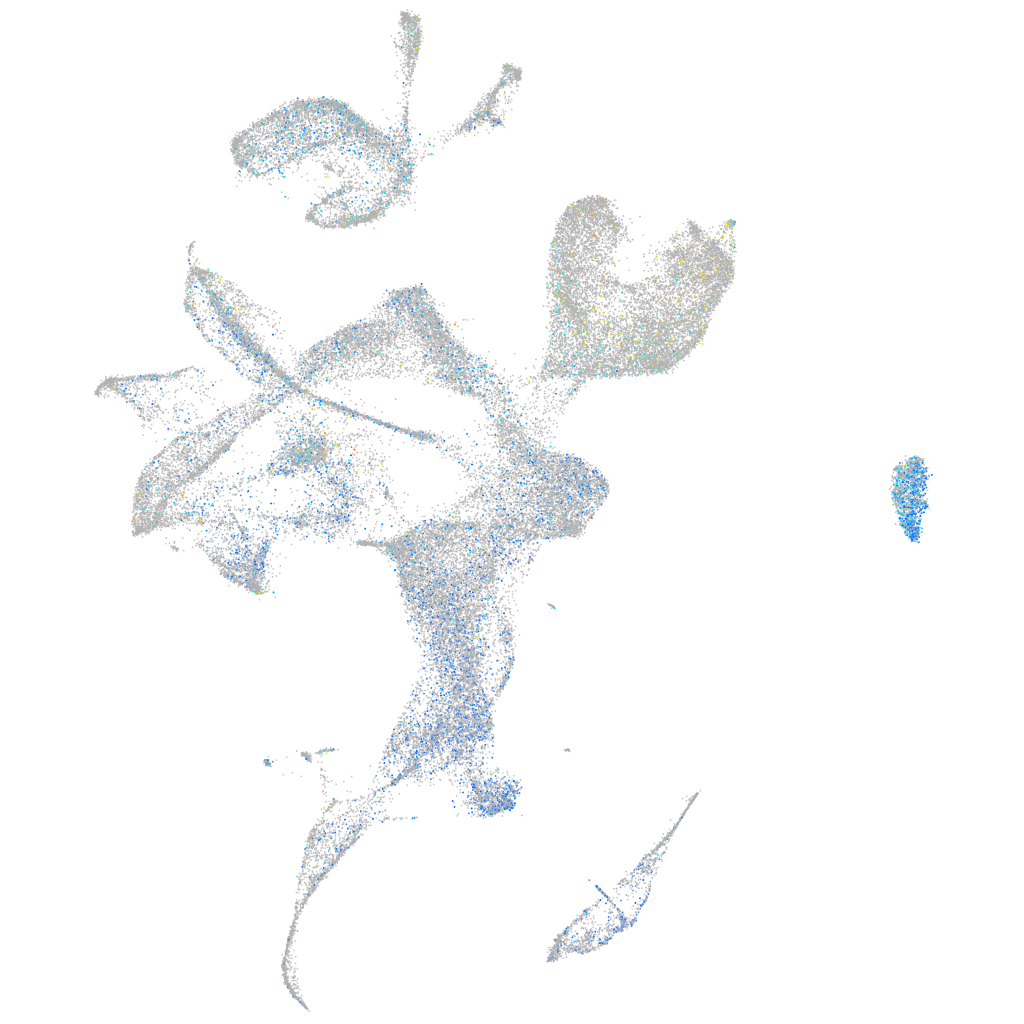
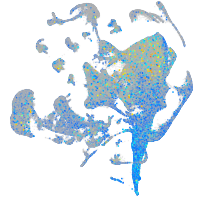
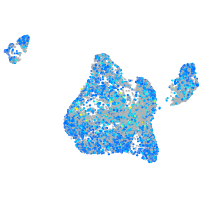
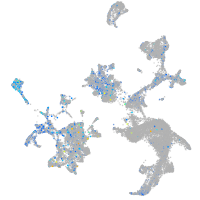
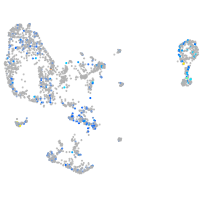
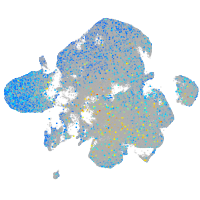
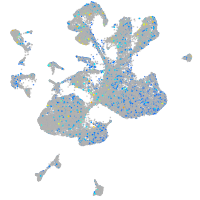
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.135 | ckbb | -0.055 |
| shisa2a | 0.120 | pvalb1 | -0.049 |
| si:ch211-152c2.3 | 0.118 | pvalb2 | -0.049 |
| stm | 0.116 | actc1b | -0.042 |
| apoeb | 0.114 | CR383676.1 | -0.041 |
| sox19a | 0.111 | mylpfa | -0.040 |
| cyp26a1 | 0.111 | rpl37 | -0.039 |
| s100a1 | 0.110 | rnasekb | -0.038 |
| zmp:0000000624 | 0.108 | syt5b | -0.037 |
| pprc1 | 0.106 | mylz3 | -0.037 |
| fbl | 0.106 | si:ch1073-429i10.3.1 | -0.037 |
| hesx1 | 0.105 | zgc:114188 | -0.037 |
| aldob | 0.104 | snap25b | -0.037 |
| nop58 | 0.104 | rbp4l | -0.036 |
| npm1a | 0.103 | rps10 | -0.036 |
| LOC108190024 | 0.103 | atpv0e2 | -0.036 |
| polr3gla | 0.101 | COX3 | -0.035 |
| dkc1 | 0.101 | mt-atp6 | -0.035 |
| asb11 | 0.101 | fabp7a | -0.035 |
| gar1 | 0.100 | ndrg1b | -0.035 |
| tdgf1 | 0.100 | ptmaa | -0.035 |
| alcamb | 0.098 | gapdhs | -0.034 |
| nr6a1a | 0.098 | rs1a | -0.033 |
| nop2 | 0.098 | neurod1 | -0.033 |
| COX7A2 | 0.098 | otx5 | -0.032 |
| nop56 | 0.097 | h3f3c | -0.032 |
| gnl3 | 0.097 | ppiab | -0.032 |
| bms1 | 0.097 | cplx4a | -0.032 |
| NC-002333.4 | 0.096 | sypb | -0.031 |
| ncl | 0.096 | h3f3a | -0.031 |
| foxd5 | 0.096 | ckmt2a | -0.031 |
| apela | 0.093 | slc25a3a | -0.031 |
| mybbp1a | 0.093 | cyt1 | -0.031 |
| pes | 0.092 | gngt2b | -0.030 |
| rrp1 | 0.092 | histh1l | -0.030 |