proline-serine-threonine phosphatase interacting protein 1b
ZFIN 
Other cell groups


all cells |
blastomeres |
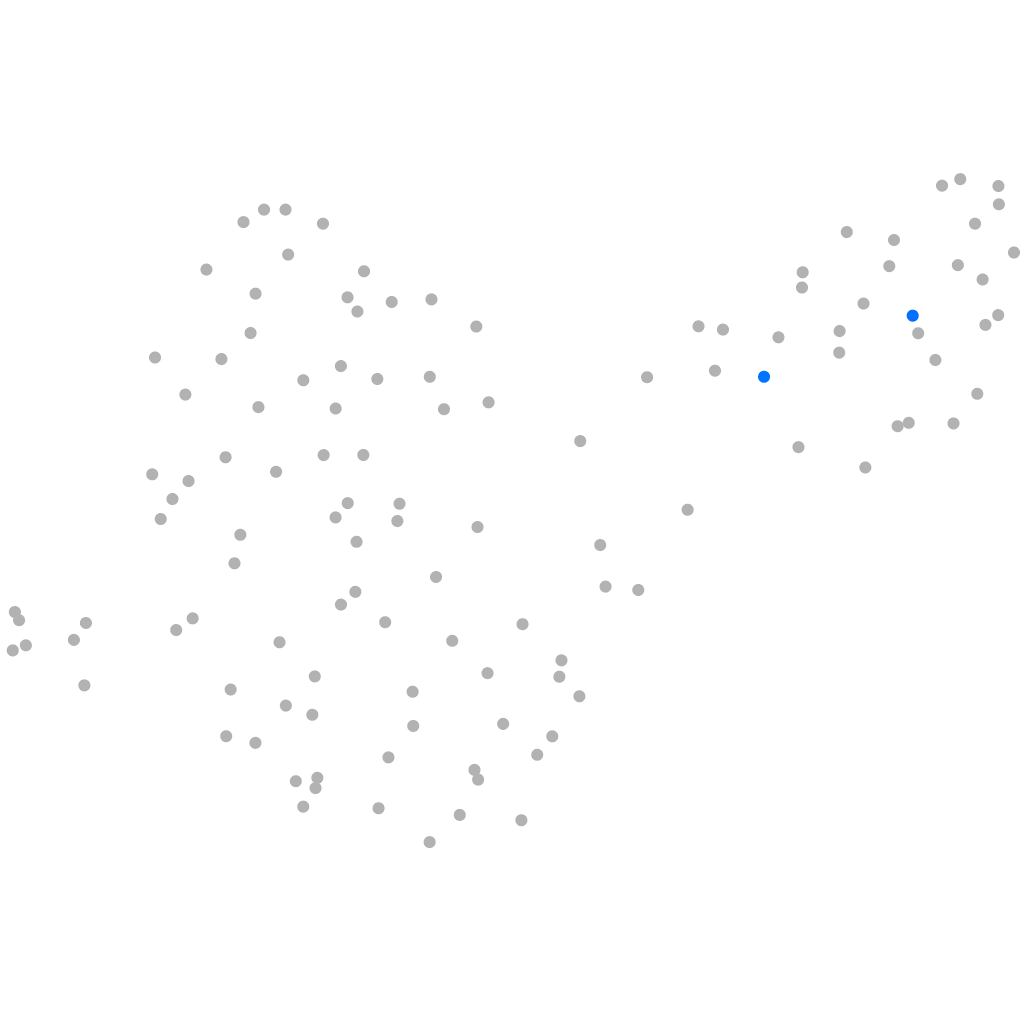
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| crispld1a | 1.000 | ybx1 | -0.174 |
| ncam1b | 0.929 | brd3a | -0.174 |
| ptprz1a | 0.759 | nucks1a | -0.172 |
| duox | 0.740 | si:ch211-214j24.10 | -0.170 |
| BX897691.1 | 0.722 | srrm1 | -0.168 |
| BX936364.1 | 0.722 | hnrnpa0a | -0.168 |
| camk1ga | 0.722 | fbl | -0.167 |
| chia.2 | 0.722 | ube2g1a | -0.165 |
| cntn3a.1 | 0.722 | marcksb | -0.164 |
| cntn3b | 0.722 | gar1 | -0.163 |
| CR318673.1 | 0.722 | larp7 | -0.158 |
| CR848746.1 | 0.722 | csnk1a1 | -0.154 |
| cryba2a | 0.722 | acin1b | -0.151 |
| crybb1 | 0.722 | htatsf1 | -0.150 |
| ctgfa | 0.722 | serbp1a | -0.149 |
| gabrr1 | 0.722 | hnrnpaba | -0.148 |
| guca1b | 0.722 | dynll2a | -0.147 |
| hcn4 | 0.722 | ncl | -0.145 |
| her13 | 0.722 | hspa9 | -0.145 |
| jdp2a | 0.722 | metap2b | -0.144 |
| LOC100151570 | 0.722 | dek | -0.144 |
| LOC101884613 | 0.722 | zgc:162025 | -0.143 |
| LOC101884995 | 0.722 | adssl | -0.141 |
| LOC108179959 | 0.722 | ddx23 | -0.140 |
| LOC562053 | 0.722 | setb | -0.140 |
| lrch2 | 0.722 | csde1 | -0.140 |
| matn4 | 0.722 | banf1 | -0.140 |
| mir203a | 0.722 | rbm7 | -0.139 |
| mxtx2 | 0.722 | tfdp1a | -0.136 |
| ngfra | 0.722 | psme3 | -0.136 |
| npy8br | 0.722 | exosc10 | -0.136 |
| parvab | 0.722 | fbxl14a | -0.136 |
| pclaf | 0.722 | ppil1 | -0.136 |
| prf1.9 | 0.722 | waca | -0.134 |
| ptpn20 | 0.722 | mphosph10 | -0.133 |