PSMC3 interacting protein
ZFIN 
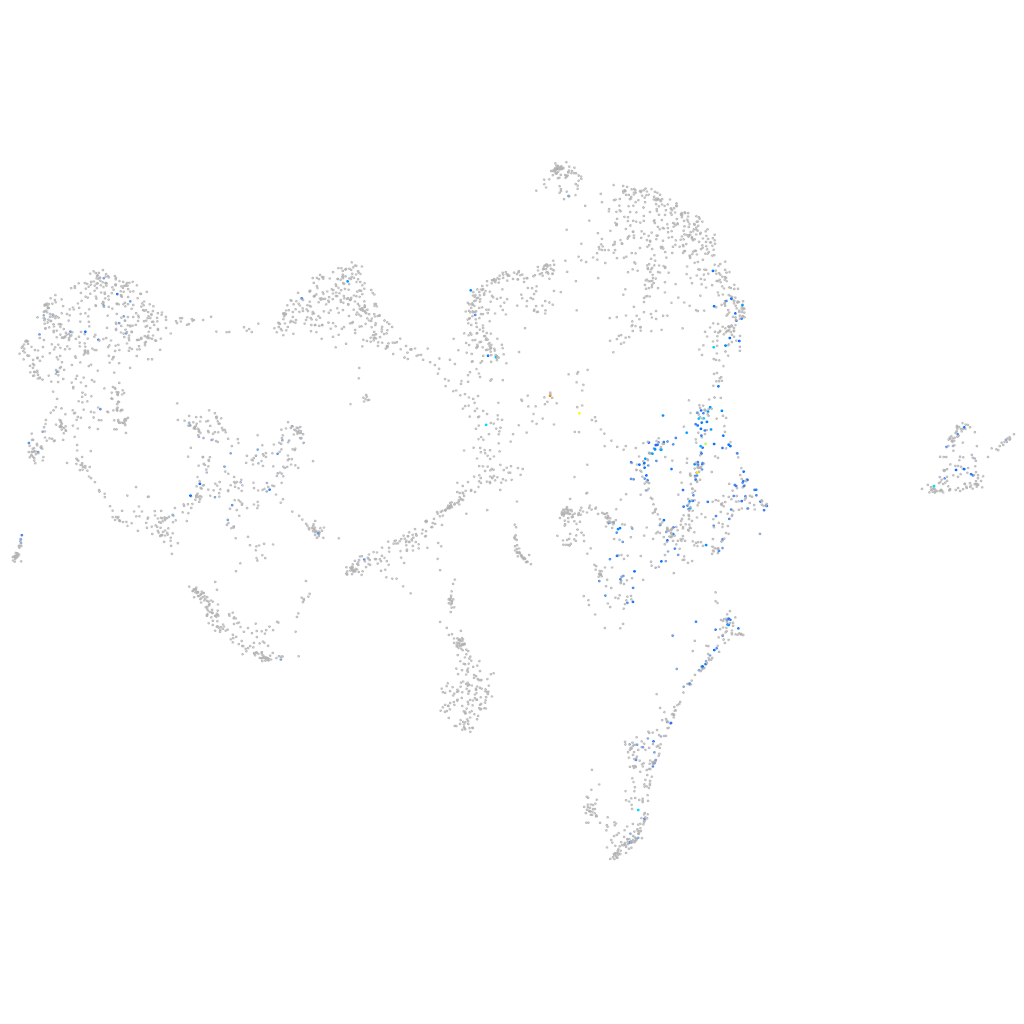
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rrm2 | 0.366 | tob1b | -0.163 |
| pcna | 0.348 | mt-co2 | -0.151 |
| rpa2 | 0.344 | si:dkey-193p11.2 | -0.149 |
| stmn1a | 0.330 | BX000438.2 | -0.149 |
| rbbp4 | 0.326 | nfe2l2a | -0.147 |
| esco2 | 0.325 | COX3 | -0.140 |
| dut | 0.325 | atp1b1b | -0.139 |
| nasp | 0.324 | selenow1 | -0.137 |
| mcm6 | 0.313 | soul2 | -0.137 |
| slbp | 0.311 | si:ch211-195b11.3 | -0.136 |
| chaf1a | 0.309 | ppdpfb | -0.132 |
| rrm1 | 0.309 | mpc1 | -0.131 |
| zgc:110540 | 0.308 | nupr1a | -0.131 |
| mcm7 | 0.300 | selenow2b | -0.129 |
| lig1 | 0.299 | tdh | -0.128 |
| rfc3 | 0.298 | mt-nd4 | -0.127 |
| tk1 | 0.297 | si:dkey-247k7.2 | -0.123 |
| fen1 | 0.296 | si:dkey-248g15.3 | -0.123 |
| rpa3 | 0.294 | mt-co1 | -0.122 |
| atad5a | 0.291 | abcb5 | -0.122 |
| hmgb2b | 0.286 | dusp2 | -0.121 |
| chek1 | 0.283 | prdx5 | -0.119 |
| ssrp1a | 0.282 | epdl1 | -0.119 |
| hmga1a | 0.274 | zgc:110333 | -0.118 |
| mibp | 0.273 | ccni | -0.117 |
| asf1ba | 0.272 | mdh2 | -0.117 |
| LOC100330864 | 0.272 | si:ch73-31d8.2 | -0.115 |
| CABZ01005379.1 | 0.269 | mt-cyb | -0.115 |
| cbx5 | 0.268 | enosf1 | -0.114 |
| prim2 | 0.267 | sqstm1 | -0.113 |
| prim1 | 0.266 | mgst1.2 | -0.112 |
| mcm2 | 0.265 | zgc:153284 | -0.111 |
| si:ch211-222l21.1 | 0.265 | bloc1s6 | -0.111 |
| sumo3b | 0.265 | sgk2a | -0.110 |
| rfc4 | 0.264 | atp6v1h | -0.109 |