"presenilin enhancer, gamma-secretase subunit"
ZFIN 
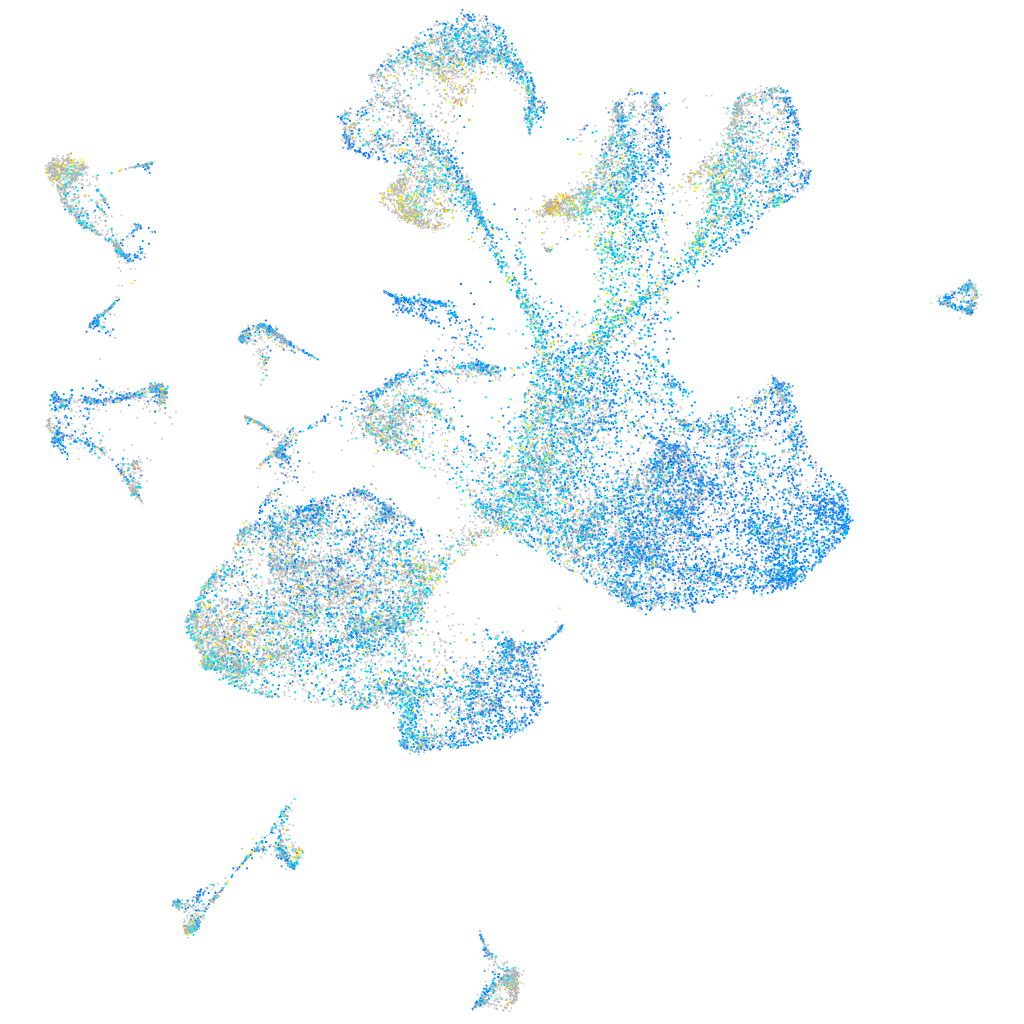
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpa0l | 0.149 | zgc:158463 | -0.074 |
| hnrnpaba | 0.141 | atp1a1b | -0.070 |
| dlb | 0.137 | gapdhs | -0.060 |
| rtca | 0.132 | fabp7a | -0.059 |
| khdrbs1a | 0.131 | eno1a | -0.056 |
| hmgn6 | 0.126 | slc1a2b | -0.054 |
| cirbpb | 0.126 | aldocb | -0.050 |
| ybx1 | 0.124 | calm1b | -0.050 |
| smarce1 | 0.124 | pvalb2 | -0.049 |
| hes6 | 0.123 | atp2b3a | -0.048 |
| hdac1 | 0.122 | sncgb | -0.048 |
| tmeff1b | 0.122 | actc1b | -0.048 |
| tp53inp2 | 0.121 | si:ch73-119p20.1 | -0.047 |
| sumo3a | 0.119 | gngt2b | -0.047 |
| si:ch211-222l21.1 | 0.119 | apoa2 | -0.046 |
| ebf2 | 0.119 | prss59.2 | -0.046 |
| hsp90ab1 | 0.118 | ckmt1 | -0.046 |
| hnrnpabb | 0.116 | vsnl1b | -0.046 |
| nhlh2 | 0.116 | elavl4 | -0.045 |
| slc25a5 | 0.115 | tpi1b | -0.045 |
| sox11b | 0.114 | pabpc1b | -0.045 |
| ubb | 0.113 | ndufa4 | -0.045 |
| gdf11 | 0.112 | eno2 | -0.045 |
| cx43.4 | 0.109 | si:dkey-16p21.8 | -0.044 |
| hnrnpa0a | 0.109 | syngr1a | -0.044 |
| robo3 | 0.107 | atp1b3b | -0.044 |
| eef1g | 0.107 | pcbp3 | -0.043 |
| snrpd2 | 0.107 | ctrb1 | -0.043 |
| ilf2 | 0.106 | pvalb1 | -0.043 |
| hnrnph1l | 0.106 | ptn | -0.043 |
| h3f3d | 0.105 | atp2b2 | -0.043 |
| cct3 | 0.104 | apoa1b | -0.043 |
| erh | 0.104 | ptprga | -0.043 |
| spsb4a | 0.103 | cavin2a | -0.042 |
| psmb7 | 0.103 | calb2b | -0.042 |