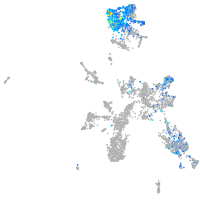
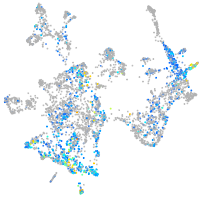
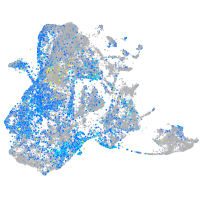
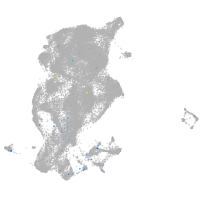
serine protease 23
ZFIN 
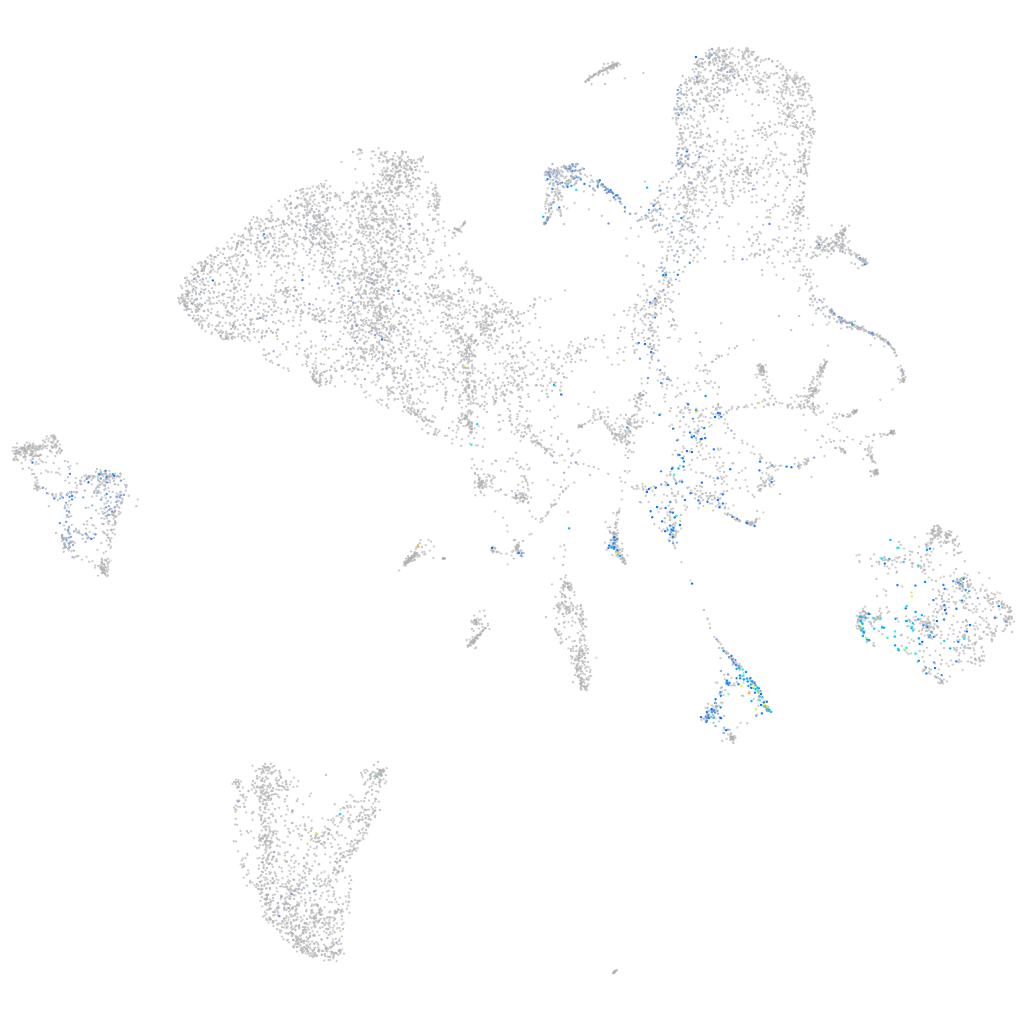
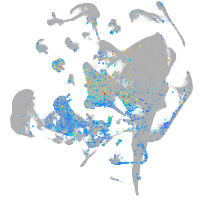
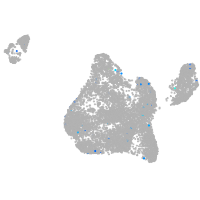
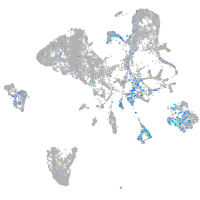
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01055365.1 | 0.309 | gapdh | -0.185 |
| ctgfa | 0.297 | ahcy | -0.183 |
| bcam | 0.286 | gatm | -0.179 |
| anxa3b | 0.264 | gamt | -0.178 |
| cd9b | 0.259 | bhmt | -0.178 |
| sim1b | 0.259 | mat1a | -0.160 |
| b3gnt5a | 0.250 | fbp1b | -0.158 |
| tgfb3 | 0.248 | apoa1b | -0.157 |
| lama5 | 0.246 | gpx4a | -0.153 |
| cabp2b | 0.245 | mdh1aa | -0.151 |
| abca12 | 0.245 | apoa4b.1 | -0.148 |
| pmp22b | 0.244 | dap | -0.147 |
| lamb1a | 0.242 | apoc2 | -0.145 |
| col4a6 | 0.241 | grhprb | -0.145 |
| mxra8b | 0.241 | apoa2 | -0.144 |
| stm | 0.237 | suclg1 | -0.143 |
| akap12b | 0.235 | agxtb | -0.143 |
| sftpba | 0.235 | afp4 | -0.143 |
| lamc1 | 0.234 | scp2a | -0.140 |
| f3a | 0.232 | agxta | -0.138 |
| gna15.1 | 0.231 | sod1 | -0.138 |
| tekt3 | 0.231 | aldh6a1 | -0.137 |
| spns2 | 0.228 | pklr | -0.137 |
| cav1 | 0.228 | abat | -0.136 |
| col4a5 | 0.222 | atp5l | -0.136 |
| tpm4a | 0.221 | gpd1b | -0.135 |
| pnp4a | 0.217 | cox6a1 | -0.134 |
| ahnak | 0.217 | aldh7a1 | -0.134 |
| msna | 0.216 | pnp4b | -0.133 |
| sox2 | 0.216 | tpi1b | -0.133 |
| spock3 | 0.216 | aqp12 | -0.132 |
| sparc | 0.215 | kng1 | -0.131 |
| CU984579.1 | 0.215 | eno3 | -0.130 |
| krt94 | 0.215 | atp5mc1 | -0.130 |
| si:ch211-264f5.6 | 0.213 | tdo2a | -0.130 |