proline rich 11
ZFIN 
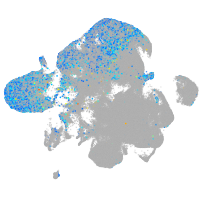
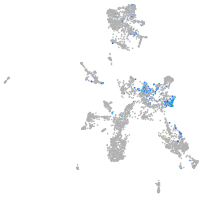
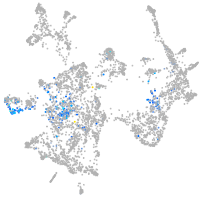
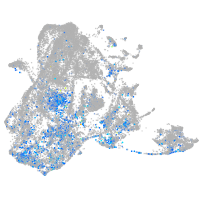
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tpx2 | 0.376 | ptmaa | -0.201 |
| ccnb1 | 0.369 | rtn1a | -0.199 |
| ube2c | 0.368 | stmn1b | -0.196 |
| mki67 | 0.359 | gng3 | -0.181 |
| plk1 | 0.356 | gpm6ab | -0.179 |
| nusap1 | 0.356 | ckbb | -0.176 |
| cdc20 | 0.355 | elavl3 | -0.175 |
| si:ch211-69g19.2 | 0.349 | sncb | -0.173 |
| mad2l1 | 0.344 | gpm6aa | -0.170 |
| cdk1 | 0.343 | tuba1c | -0.169 |
| kpna2 | 0.336 | tmsb | -0.167 |
| cenpf | 0.335 | rnasekb | -0.165 |
| cdca8 | 0.332 | rtn1b | -0.164 |
| aurka | 0.332 | atp6v0cb | -0.163 |
| kif11 | 0.330 | ywhah | -0.156 |
| aspm | 0.330 | vamp2 | -0.154 |
| aurkb | 0.329 | elavl4 | -0.154 |
| prc1a | 0.326 | atp6v1e1b | -0.153 |
| top2a | 0.323 | ywhag2 | -0.151 |
| kmt5ab | 0.320 | fez1 | -0.150 |
| kifc1 | 0.318 | calm1a | -0.150 |
| kif23 | 0.312 | stmn2a | -0.148 |
| ccna2 | 0.302 | COX3 | -0.147 |
| dlgap5 | 0.299 | stx1b | -0.146 |
| ccnb3 | 0.295 | gapdhs | -0.146 |
| tacc3 | 0.295 | tubb5 | -0.146 |
| birc5a | 0.295 | stxbp1a | -0.143 |
| armc1l | 0.287 | dpysl3 | -0.142 |
| g2e3 | 0.283 | kdm6bb | -0.141 |
| cenpe | 0.282 | si:dkeyp-75h12.5 | -0.141 |
| pif1 | 0.281 | gap43 | -0.141 |
| spdl1 | 0.279 | mllt11 | -0.140 |
| hmgb2a | 0.277 | atp6v1g1 | -0.140 |
| kif20a | 0.277 | zgc:65894 | -0.139 |
| kif22 | 0.276 | pvalb1 | -0.139 |