"peripherin 2, like [Source:ZFIN;Acc:ZDB-GENE-041001-147]"
ZFIN 
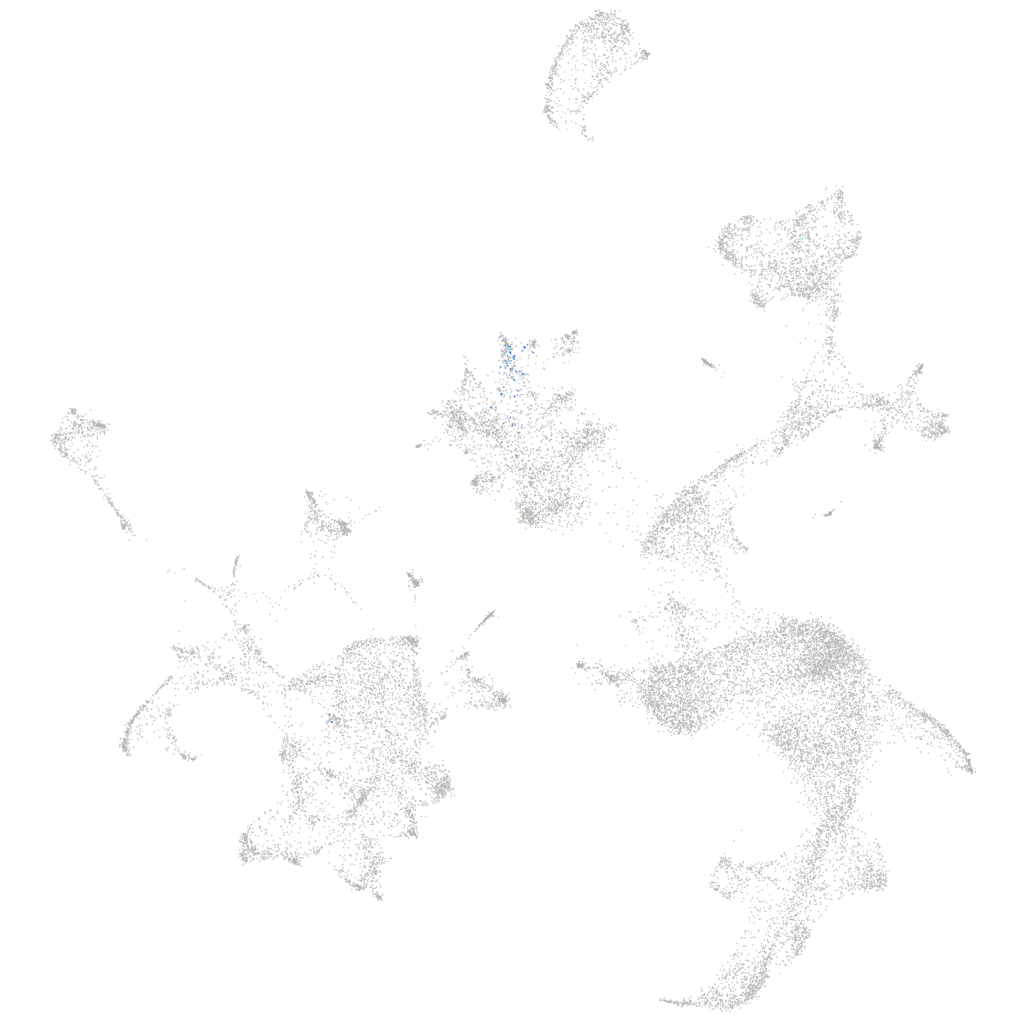
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm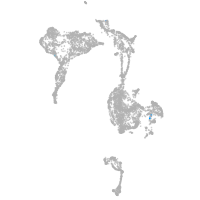 |
muscle  |
mural cells / non-skeletal muscle 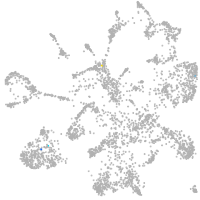 |
mesenchyme  |
hematopoietic / vasculature 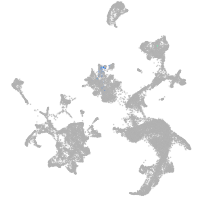 |
pronephros  |
neural |
spinal cord / glia |
eye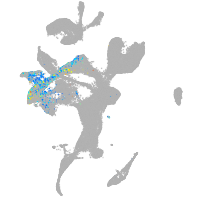 |
taste / olfactory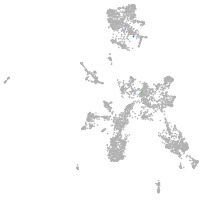 |
otic / lateral line |
epidermis |
periderm |
fin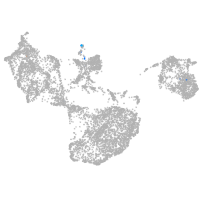 |
ionocytes / mucous |
pigment cells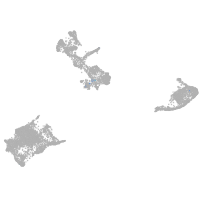 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| guk1b | 0.288 | arl4aa | -0.018 |
| gngt2a | 0.288 | rpl9 | -0.017 |
| arl3l1 | 0.283 | hbbe3 | -0.017 |
| rx2 | 0.281 | rhocb | -0.017 |
| arr3a | 0.279 | zgc:56493 | -0.016 |
| rcvrn2 | 0.277 | yrk | -0.016 |
| zgc:103625 | 0.272 | pvalb1 | -0.016 |
| kcnb2 | 0.267 | ehd4 | -0.016 |
| glulc | 0.266 | ucp2 | -0.016 |
| rbp4l | 0.266 | clic2 | -0.016 |
| atp2b1b | 0.265 | mcl1a | -0.015 |
| cnga3b | 0.262 | si:dkey-112e7.2 | -0.015 |
| si:ch1073-469d17.2 | 0.261 | CT030188.1 | -0.015 |
| arl3l2 | 0.258 | znfl2a | -0.015 |
| tsga10 | 0.258 | glrx | -0.015 |
| cngb3.2 | 0.251 | lmo2 | -0.015 |
| faimb | 0.248 | cdh5 | -0.015 |
| ckmt2a | 0.248 | aqp1a.1 | -0.015 |
| si:dkey-220f10.4 | 0.246 | plvapb | -0.015 |
| arl13a | 0.246 | kdr | -0.015 |
| reep6 | 0.244 | etv2 | -0.015 |
| tulp1a | 0.241 | ecscr | -0.014 |
| tmem244 | 0.240 | myct1a | -0.014 |
| zgc:109965 | 0.239 | bzw1b | -0.014 |
| opn6a | 0.237 | actc1b | -0.014 |
| tmx3a | 0.236 | srgn | -0.014 |
| rcvrna | 0.236 | mfng | -0.014 |
| elovl4b | 0.234 | si:ch73-248e21.7 | -0.014 |
| slc25a3a | 0.233 | mcamb | -0.014 |
| si:ch211-81a5.8 | 0.232 | tal1 | -0.014 |
| pdcb | 0.230 | banf1 | -0.014 |
| prph2a | 0.229 | csrp1a | -0.014 |
| inaa | 0.229 | clec14a | -0.014 |
| si:dkey-26i13.8 | 0.226 | pfn1 | -0.014 |
| LOC103909376 | 0.226 | sgk1 | -0.014 |